Please be patient as the page loads
|
AKAP8_HUMAN
|
||||||
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AKAP8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 166 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AKAP8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.958300 (rank : 2) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AKP8L_MOUSE
|
||||||
| θ value | 3.58603e-65 (rank : 3) | NC score | 0.854676 (rank : 4) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0L7 | Gene names | Akap8l, Nakap95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95). | |||||
|
AKP8L_HUMAN
|
||||||
| θ value | 1.15359e-63 (rank : 4) | NC score | 0.857550 (rank : 3) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 8.91499e-32 (rank : 5) | NC score | 0.723023 (rank : 5) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
ZN326_HUMAN
|
||||||
| θ value | 3.17079e-29 (rank : 6) | NC score | 0.716624 (rank : 6) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 7) | NC score | 0.082997 (rank : 13) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 8) | NC score | 0.145470 (rank : 7) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 9) | NC score | 0.076042 (rank : 17) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.025176 (rank : 102) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.076694 (rank : 15) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 12) | NC score | 0.050395 (rank : 46) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 13) | NC score | 0.081506 (rank : 14) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.036565 (rank : 76) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
ZFX_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.020590 (rank : 116) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17012, P17011 | Gene names | Zfx | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger X-chromosomal protein. | |||||
|
ZFY_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.020964 (rank : 114) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08048, Q14021, Q15558, Q96TF3 | Gene names | ZFY | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Y-chromosomal protein. | |||||
|
AN32A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.056779 (rank : 30) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P39687 | Gene names | ANP32A, C15orf1, LANP, MAPM, PHAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein) (Lanp) (Putative HLA-DR-associated protein I) (PHAPI) (Mapmodulin). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.092013 (rank : 8) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TB182_HUMAN
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.083952 (rank : 12) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
ZFA_MOUSE
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.020697 (rank : 115) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P23607 | Gene names | Zfa | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger autosomal protein. | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.085501 (rank : 11) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MFA3L_MOUSE
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.032695 (rank : 83) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D3X9, Q80TV6, Q8BJA9 | Gene names | Mfap3l, Kiaa0626 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microfibrillar-associated protein 3-like precursor. | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.056234 (rank : 31) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ZFX_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.020124 (rank : 117) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17010, O43668, Q8WYJ8 | Gene names | ZFX | |||
|
Domain Architecture |
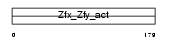
|
|||||
| Description | Zinc finger X-chromosomal protein. | |||||
|
A4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.056136 (rank : 32) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.051998 (rank : 42) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.016414 (rank : 136) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
PALM_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.050452 (rank : 45) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
ZN131_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.013171 (rank : 149) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P52739, Q6PIF0 | Gene names | ZNF131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 131. | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.031012 (rank : 86) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
K1279_MOUSE
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.042692 (rank : 67) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPU9, Q5BKR5, Q7TPE0, Q80VQ5, Q8BZE2, Q8CFS7 | Gene names | Kiaa1279 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1279. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.088233 (rank : 10) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
AN32A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.055917 (rank : 33) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.055917 (rank : 34) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
IQEC3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.023609 (rank : 106) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPP2, Q8TB43 | Gene names | IQSEC3, KIAA1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
MPP10_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.059248 (rank : 28) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
TNNT3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.053063 (rank : 40) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.007279 (rank : 167) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ROA2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.050132 (rank : 48) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22626, P22627 | Gene names | HNRPA2B1 | |||
|
Domain Architecture |
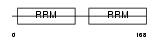
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.055055 (rank : 36) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
FBLN2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.008238 (rank : 164) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.049130 (rank : 53) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.069706 (rank : 19) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
A4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.038806 (rank : 71) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
HGFA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.004660 (rank : 173) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
IRX4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.018935 (rank : 122) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QY61 | Gene names | Irx4, Irxa3 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-4 (Iroquois homeobox protein 4) (Homeodomain protein IRXA3). | |||||
|
NHRF2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.016249 (rank : 137) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.046950 (rank : 59) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
REN3A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.054627 (rank : 37) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.038844 (rank : 70) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
AP180_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.027467 (rank : 96) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
HNRPD_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.034349 (rank : 80) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14103, Q01858, Q14100, Q14101, Q14102 | Gene names | HNRPD, AUF1 | |||
|
Domain Architecture |
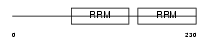
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
HNRPD_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.034418 (rank : 78) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60668, Q60667, Q80ZJ0, Q91X94 | Gene names | Hnrpd, Auf1 | |||
|
Domain Architecture |
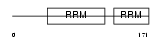
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.034537 (rank : 77) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
ILF3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.051915 (rank : 43) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.066995 (rank : 21) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.069210 (rank : 20) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.027895 (rank : 95) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.088261 (rank : 9) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PWP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.014551 (rank : 143) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LL5, Q9D6T6 | Gene names | Pwp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periodic tryptophan protein 1 homolog. | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.053688 (rank : 38) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
TRI55_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.016157 (rank : 138) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYV6, Q53XX3, Q8IUD9, Q8IUE4, Q96DV2, Q96DV3, Q9BYV5 | Gene names | TRIM55, MURF2, RNF29 | |||
|
Domain Architecture |
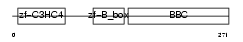
|
|||||
| Description | Tripartite motif-containing protein 55 (Muscle-specific RING finger protein 2) (MuRF2) (MURF-2) (RING finger protein 29). | |||||
|
VKGC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.048046 (rank : 55) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYC7, Q3UXN5, Q8CCB3 | Gene names | Ggcx | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent gamma-carboxylase (EC 6.4.-.-) (Gamma-glutamyl carboxylase) (Vitamin K gamma glutamyl carboxylase). | |||||
|
ZN132_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.008238 (rank : 163) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P52740 | Gene names | ZNF132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 132. | |||||
|
GAGC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.057354 (rank : 29) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60829, Q6IBI1 | Gene names | PAGE4, GAGEC1 | |||
|
Domain Architecture |
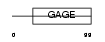
|
|||||
| Description | G antigen family C member 1 (Prostate-associated gene 4 protein) (PAGE-4) (PAGE-1) (GAGE-9). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.023543 (rank : 107) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.037811 (rank : 74) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.013506 (rank : 146) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
ROA3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.049296 (rank : 52) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BG05, Q8BHF8 | Gene names | Hnrpa3, Hnrnpa3 | |||
|
Domain Architecture |
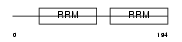
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.063163 (rank : 24) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CHMP6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.030103 (rank : 90) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.042289 (rank : 68) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
HD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.021842 (rank : 112) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.076568 (rank : 16) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KAP0_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.017233 (rank : 132) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
ROA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.045309 (rank : 63) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88569 | Gene names | Hnrpa2b1 | |||
|
Domain Architecture |
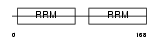
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
TNNT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.033393 (rank : 82) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P45378, Q12975, Q12976, Q12977, Q12978, Q86TH6 | Gene names | TNNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT) (Beta TnTF). | |||||
|
TROP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.013336 (rank : 147) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.018362 (rank : 125) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ASPX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.053422 (rank : 39) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.007230 (rank : 168) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
GAGE3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.050151 (rank : 47) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13067 | Gene names | GAGE3 | |||
|
Domain Architecture |
|
|||||
| Description | G antigen 3 (GAGE-3). | |||||
|
GAGE5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.049578 (rank : 51) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13069, Q6FG72 | Gene names | GAGE5 | |||
|
Domain Architecture |
|
|||||
| Description | G antigen 5 (GAGE-5). | |||||
|
GAGE6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.050065 (rank : 49) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13070 | Gene names | GAGE6 | |||
|
Domain Architecture |
|
|||||
| Description | G antigen 6 (GAGE-6). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.016878 (rank : 135) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.027131 (rank : 98) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NNP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.041213 (rank : 69) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P56183, O35712, Q9ERE1, Q9JI07, Q9JK67, Q9JKU2 | Gene names | Nnp1 | |||
|
Domain Architecture |
|
|||||
| Description | NNP-1 protein (Novel nuclear protein 1) (Nop52). | |||||
|
NOP14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.032518 (rank : 84) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
PEDF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.006772 (rank : 169) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97298, O70629, O88691 | Gene names | Serpinf1, Pedf, Sdf3 | |||
|
Domain Architecture |
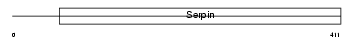
|
|||||
| Description | Pigment epithelium-derived factor precursor (PEDF) (Serpin-F1) (Stromal cell-derived factor 3) (SDF-3) (Caspin). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.008342 (rank : 162) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
SDC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.030597 (rank : 88) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P18827, Q96HB7 | Gene names | SDC1, SDC | |||
|
Domain Architecture |
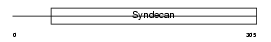
|
|||||
| Description | Syndecan-1 precursor (SYND1) (CD138 antigen). | |||||
|
SH3BG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.024907 (rank : 103) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55822, Q9BRB8 | Gene names | SH3BGR | |||
|
Domain Architecture |
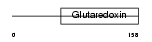
|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein) (21- glutamic acid-rich protein) (21-GARP). | |||||
|
SUHW3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.015315 (rank : 141) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8ND82, Q9NXR3 | Gene names | SUHW3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
ZFY1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.018231 (rank : 126) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
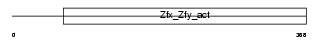
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
ZFY2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.018143 (rank : 128) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
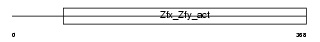
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
41_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.009793 (rank : 158) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.011026 (rank : 155) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CAMKV_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.003048 (rank : 174) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3UHL1, Q8VD20 | Gene names | Camkv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
DEK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.037749 (rank : 75) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.017183 (rank : 133) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
K1C10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.018462 (rank : 124) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
NFL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.024592 (rank : 104) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
NKX22_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.012034 (rank : 152) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95096 | Gene names | NKX2-2, NKX2.2, NKX2B | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.2 (Homeobox protein NK-2 homolog B). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.027902 (rank : 94) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.018883 (rank : 123) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.021936 (rank : 111) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
TTC15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.030442 (rank : 89) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WVT3, Q8WVW1, Q9Y395 | Gene names | TTC15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
VKGC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.042981 (rank : 66) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38435, Q14415, Q6GU45 | Gene names | GGCX, GC | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent gamma-carboxylase (EC 6.4.-.-) (Gamma-glutamyl carboxylase) (Vitamin K gamma glutamyl carboxylase). | |||||
|
Z3H7A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.024459 (rank : 105) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.014269 (rank : 144) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.038678 (rank : 72) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
DGKK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.022573 (rank : 110) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.010257 (rank : 156) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
GATA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.009737 (rank : 159) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |
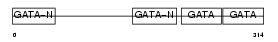
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
NECD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.012504 (rank : 150) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |
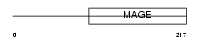
|
|||||
| Description | Necdin. | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.030732 (rank : 87) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PHX2B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.007989 (rank : 165) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99453, Q6PJD9 | Gene names | PHOX2B, PMX2B | |||
|
Domain Architecture |
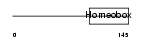
|
|||||
| Description | Paired mesoderm homeobox protein 2B (Paired-like homeobox 2B) (PHOX2B homeodomain protein) (Neuroblastoma Phox) (NBPhox). | |||||
|
PHX2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.007989 (rank : 166) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35690 | Gene names | Phox2b, Pmx2b | |||
|
Domain Architecture |
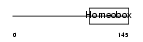
|
|||||
| Description | Paired mesoderm homeobox protein 2B (Paired-like homeobox 2B) (PHOX2B homeodomain protein) (Neuroblastoma Phox) (NBPhox). | |||||
|
PIN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.021290 (rank : 113) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.025554 (rank : 99) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
ROA0_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.045105 (rank : 64) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13151 | Gene names | HNRPA0 | |||
|
Domain Architecture |
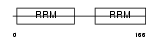
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A0 (hnRNP A0). | |||||
|
RTN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.017822 (rank : 130) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75298, O60509, Q7RTM6, Q7RTN1, Q7RTN2 | Gene names | RTN2, NSPL1 | |||
|
Domain Architecture |
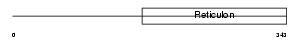
|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.061120 (rank : 27) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TAF10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.025198 (rank : 101) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K0H5, Q8C8H2, Q9QYY5 | Gene names | Taf10, Taf2h, Tafii30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 10 (Transcription initiation factor TFIID 30 kDa subunit) (TAF(II)30) (TAFII-30) (mTAFII30). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.028044 (rank : 93) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.033924 (rank : 81) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
TSH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.019799 (rank : 119) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.023532 (rank : 108) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.012499 (rank : 151) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
GAGE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.046832 (rank : 60) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13065 | Gene names | GAGE1 | |||
|
Domain Architecture |
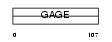
|
|||||
| Description | G antigen 1 (GAGE-1) (MZ2-F antigen). | |||||
|
GAGE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.046039 (rank : 62) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13066, Q4V322 | Gene names | GAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G antigen 2 (GAGE-2). | |||||
|
GAGE4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.046057 (rank : 61) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13068, Q6NT33 | Gene names | GAGE4 | |||
|
Domain Architecture |
|
|||||
| Description | G antigen 4 (GAGE-4). | |||||
|
GAGE7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.045012 (rank : 65) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O76087, Q4V324 | Gene names | GAGE7B, GAGE7 | |||
|
Domain Architecture |
|
|||||
| Description | G antigen 7 (GAGE-7) (GAGE-7B) (GAGE-8) (AL4). | |||||
|
GAGE8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.047080 (rank : 58) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UEU5, Q4V325 | Gene names | GAGE8 | |||
|
Domain Architecture |
|
|||||
| Description | G antigen 8 (GAGE-8). | |||||
|
GG12A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.048034 (rank : 56) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q4V321 | Gene names | GAGE12A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G antigen 12A (GAGE-12A). | |||||
|
GNAS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.009651 (rank : 161) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
IF38_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.025461 (rank : 100) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99613, O00215 | Gene names | EIF3S8 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110) (eIF3c). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.034411 (rank : 79) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.038423 (rank : 73) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.048290 (rank : 54) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
PARPT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.011723 (rank : 153) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
RBM14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.019433 (rank : 121) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
RBM14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.018177 (rank : 127) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
ROBO4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.005118 (rank : 172) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |
|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.049954 (rank : 50) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SOX3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.005592 (rank : 171) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SYNJ2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.009675 (rank : 160) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
ASXL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.015940 (rank : 139) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
CCD11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.014635 (rank : 142) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.019922 (rank : 118) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
DEK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.028603 (rank : 91) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ELMD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.010159 (rank : 157) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N336, Q9NPW3 | Gene names | ELMOD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELMO domain-containing protein 1. | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.013725 (rank : 145) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
IF35_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.019505 (rank : 120) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.013293 (rank : 148) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.022600 (rank : 109) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.047385 (rank : 57) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NF2L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.017126 (rank : 134) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.031451 (rank : 85) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PIN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.017892 (rank : 129) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
RBM19_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.017814 (rank : 131) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.028177 (rank : 92) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SDCG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.015527 (rank : 140) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |
|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.006764 (rank : 170) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
TOIP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.027299 (rank : 97) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.011480 (rank : 154) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.061820 (rank : 26) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.061960 (rank : 25) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.055369 (rank : 35) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.065397 (rank : 23) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.069974 (rank : 18) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.050542 (rank : 44) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.066926 (rank : 22) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.052386 (rank : 41) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
AKAP8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 166 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AKAP8_MOUSE
|
||||||
| NC score | 0.958300 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AKP8L_HUMAN
|
||||||
| NC score | 0.857550 (rank : 3) | θ value | 1.15359e-63 (rank : 4) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
AKP8L_MOUSE
|
||||||
| NC score | 0.854676 (rank : 4) | θ value | 3.58603e-65 (rank : 3) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0L7 | Gene names | Akap8l, Nakap95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95). | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.723023 (rank : 5) | θ value | 8.91499e-32 (rank : 5) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
ZN326_HUMAN
|
||||||
| NC score | 0.716624 (rank : 6) | θ value | 3.17079e-29 (rank : 6) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.145470 (rank : 7) | θ value | 0.00228821 (rank : 8) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.092013 (rank : 8) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.088261 (rank : 9) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.088233 (rank : 10) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.085501 (rank : 11) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.083952 (rank : 12) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.082997 (rank : 13) | θ value | 0.000602161 (rank : 7) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.081506 (rank : 14) | θ value | 0.0431538 (rank : 13) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.076694 (rank : 15) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.076568 (rank : 16) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.076042 (rank : 17) | θ value | 0.00665767 (rank : 9) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.069974 (rank : 18) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.069706 (rank : 19) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.069210 (rank : 20) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.066995 (rank : 21) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.066926 (rank : 22) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.065397 (rank : 23) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.063163 (rank : 24) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.061960 (rank : 25) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.061820 (rank : 26) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.061120 (rank : 27) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
MPP10_HUMAN
|
||||||
| NC score | 0.059248 (rank : 28) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
GAGC1_HUMAN
|
||||||
| NC score | 0.057354 (rank : 29) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60829, Q6IBI1 | Gene names | PAGE4, GAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen family C member 1 (Prostate-associated gene 4 protein) (PAGE-4) (PAGE-1) (GAGE-9). | |||||
|
AN32A_HUMAN
|
||||||
| NC score | 0.056779 (rank : 30) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P39687 | Gene names | ANP32A, C15orf1, LANP, MAPM, PHAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein) (Lanp) (Putative HLA-DR-associated protein I) (PHAPI) (Mapmodulin). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.056234 (rank : 31) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.056136 (rank : 32) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
AN32A_MOUSE
|
||||||
| NC score | 0.055917 (rank : 33) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| NC score | 0.055917 (rank : 34) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.055369 (rank : 35) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.055055 (rank : 36) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
REN3A_HUMAN
|
||||||
| NC score | 0.054627 (rank : 37) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.053688 (rank : 38) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
ASPX_HUMAN
|
||||||
| NC score | 0.053422 (rank : 39) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
TNNT3_MOUSE
|
||||||
| NC score | 0.053063 (rank : 40) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.052386 (rank : 41) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.051998 (rank : 42) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ILF3_HUMAN
|
||||||
| NC score | 0.051915 (rank : 43) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.050542 (rank : 44) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PALM_MOUSE
|
||||||
| NC score | 0.050452 (rank : 45) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.050395 (rank : 46) | θ value | 0.0431538 (rank : 12) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
GAGE3_HUMAN
|
||||||
| NC score | 0.050151 (rank : 47) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13067 | Gene names | GAGE3 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen 3 (GAGE-3). | |||||
|
ROA2_HUMAN
|
||||||
| NC score | 0.050132 (rank : 48) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22626, P22627 | Gene names | HNRPA2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
GAGE6_HUMAN
|
||||||
| NC score | 0.050065 (rank : 49) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13070 | Gene names | GAGE6 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen 6 (GAGE-6). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.049954 (rank : 50) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
GAGE5_HUMAN
|
||||||
| NC score | 0.049578 (rank : 51) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13069, Q6FG72 | Gene names | GAGE5 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen 5 (GAGE-5). | |||||
|
ROA3_MOUSE
|
||||||
| NC score | 0.049296 (rank : 52) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BG05, Q8BHF8 | Gene names | Hnrpa3, Hnrnpa3 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.049130 (rank : 53) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.048290 (rank : 54) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
VKGC_MOUSE
|
||||||
| NC score | 0.048046 (rank : 55) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYC7, Q3UXN5, Q8CCB3 | Gene names | Ggcx | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent gamma-carboxylase (EC 6.4.-.-) (Gamma-glutamyl carboxylase) (Vitamin K gamma glutamyl carboxylase). | |||||
|
GG12A_HUMAN
|
||||||
| NC score | 0.048034 (rank : 56) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q4V321 | Gene names | GAGE12A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G antigen 12A (GAGE-12A). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.047385 (rank : 57) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
GAGE8_HUMAN
|
||||||
| NC score | 0.047080 (rank : 58) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UEU5, Q4V325 | Gene names | GAGE8 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen 8 (GAGE-8). | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.046950 (rank : 59) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
GAGE1_HUMAN
|
||||||
| NC score | 0.046832 (rank : 60) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13065 | Gene names | GAGE1 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen 1 (GAGE-1) (MZ2-F antigen). | |||||
|
GAGE4_HUMAN
|
||||||
| NC score | 0.046057 (rank : 61) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13068, Q6NT33 | Gene names | GAGE4 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen 4 (GAGE-4). | |||||
|
GAGE2_HUMAN
|
||||||
| NC score | 0.046039 (rank : 62) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13066, Q4V322 | Gene names | GAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G antigen 2 (GAGE-2). | |||||
|
ROA2_MOUSE
|
||||||
| NC score | 0.045309 (rank : 63) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88569 | Gene names | Hnrpa2b1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
ROA0_HUMAN
|
||||||
| NC score | 0.045105 (rank : 64) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13151 | Gene names | HNRPA0 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A0 (hnRNP A0). | |||||
|
GAGE7_HUMAN
|
||||||
| NC score | 0.045012 (rank : 65) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O76087, Q4V324 | Gene names | GAGE7B, GAGE7 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen 7 (GAGE-7) (GAGE-7B) (GAGE-8) (AL4). | |||||
|
VKGC_HUMAN
|
||||||
| NC score | 0.042981 (rank : 66) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38435, Q14415, Q6GU45 | Gene names | GGCX, GC | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent gamma-carboxylase (EC 6.4.-.-) (Gamma-glutamyl carboxylase) (Vitamin K gamma glutamyl carboxylase). | |||||
|
K1279_MOUSE
|
||||||
| NC score | 0.042692 (rank : 67) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPU9, Q5BKR5, Q7TPE0, Q80VQ5, Q8BZE2, Q8CFS7 | Gene names | Kiaa1279 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1279. | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.042289 (rank : 68) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
NNP1_MOUSE
|
||||||
| NC score | 0.041213 (rank : 69) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P56183, O35712, Q9ERE1, Q9JI07, Q9JK67, Q9JKU2 | Gene names | Nnp1 | |||
|
Domain Architecture |

|
|||||
| Description | NNP-1 protein (Novel nuclear protein 1) (Nop52). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.038844 (rank : 70) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.038806 (rank : 71) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.038678 (rank : 72) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.038423 (rank : 73) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.037811 (rank : 74) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.037749 (rank : 75) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.036565 (rank : 76) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.034537 (rank : 77) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
HNRPD_MOUSE
|
||||||
| NC score | 0.034418 (rank : 78) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60668, Q60667, Q80ZJ0, Q91X94 | Gene names | Hnrpd, Auf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.034411 (rank : 79) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
HNRPD_HUMAN
|
||||||
| NC score | 0.034349 (rank : 80) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14103, Q01858, Q14100, Q14101, Q14102 | Gene names | HNRPD, AUF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.033924 (rank : 81) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
TNNT3_HUMAN
|
||||||
| NC score | 0.033393 (rank : 82) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P45378, Q12975, Q12976, Q12977, Q12978, Q86TH6 | Gene names | TNNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT) (Beta TnTF). | |||||
|
MFA3L_MOUSE
|
||||||
| NC score | 0.032695 (rank : 83) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D3X9, Q80TV6, Q8BJA9 | Gene names | Mfap3l, Kiaa0626 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microfibrillar-associated protein 3-like precursor. | |||||
|
NOP14_MOUSE
|
||||||
| NC score | 0.032518 (rank : 84) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.031451 (rank : 85) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.031012 (rank : 86) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.030732 (rank : 87) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SDC1_HUMAN
|
||||||
| NC score | 0.030597 (rank : 88) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P18827, Q96HB7 | Gene names | SDC1, SDC | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-1 precursor (SYND1) (CD138 antigen). | |||||
|
TTC15_HUMAN
|
||||||
| NC score | 0.030442 (rank : 89) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WVT3, Q8WVW1, Q9Y395 | Gene names | TTC15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
CHMP6_MOUSE
|
||||||
| NC score | 0.030103 (rank : 90) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.028603 (rank : 91) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.028177 (rank : 92) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.028044 (rank : 93) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.027902 (rank : 94) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.027895 (rank : 95) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
AP180_MOUSE
|
||||||
| NC score | 0.027467 (rank : 96) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
TOIP2_MOUSE
|
||||||
| NC score | 0.027299 (rank : 97) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.027131 (rank : 98) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.025554 (rank : 99) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
IF38_HUMAN
|
||||||
| NC score | 0.025461 (rank : 100) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99613, O00215 | Gene names | EIF3S8 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110) (eIF3c). | |||||
|
TAF10_MOUSE
|
||||||
| NC score | 0.025198 (rank : 101) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K0H5, Q8C8H2, Q9QYY5 | Gene names | Taf10, Taf2h, Tafii30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 10 (Transcription initiation factor TFIID 30 kDa subunit) (TAF(II)30) (TAFII-30) (mTAFII30). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.025176 (rank : 102) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
SH3BG_HUMAN
|
||||||
| NC score | 0.024907 (rank : 103) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55822, Q9BRB8 | Gene names | SH3BGR | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein) (21- glutamic acid-rich protein) (21-GARP). | |||||
|
NFL_HUMAN
|
||||||
| NC score | 0.024592 (rank : 104) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
Z3H7A_HUMAN
|
||||||
| NC score | 0.024459 (rank : 105) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
IQEC3_HUMAN
|
||||||
| NC score | 0.023609 (rank : 106) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPP2, Q8TB43 | Gene names | IQSEC3, KIAA1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.023543 (rank : 107) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.023532 (rank : 108) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.022600 (rank : 109) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.022573 (rank : 110) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.021936 (rank : 111) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.021842 (rank : 112) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
PIN1_HUMAN
|
||||||
| NC score | 0.021290 (rank : 113) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
ZFY_HUMAN
|
||||||
| NC score | 0.020964 (rank : 114) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08048, Q14021, Q15558, Q96TF3 | Gene names | ZFY | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Y-chromosomal protein. | |||||
|
ZFA_MOUSE
|
||||||
| NC score | 0.020697 (rank : 115) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P23607 | Gene names | Zfa | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger autosomal protein. | |||||
|
ZFX_MOUSE
|
||||||
| NC score | 0.020590 (rank : 116) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17012, P17011 | Gene names | Zfx | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger X-chromosomal protein. | |||||
|
ZFX_HUMAN
|
||||||
| NC score | 0.020124 (rank : 117) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17010, O43668, Q8WYJ8 | Gene names | ZFX | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger X-chromosomal protein. | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.019922 (rank : 118) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
TSH1_HUMAN
|
||||||
| NC score | 0.019799 (rank : 119) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.019505 (rank : 120) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
RBM14_HUMAN
|
||||||
| NC score | 0.019433 (rank : 121) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
IRX4_MOUSE
|
||||||
| NC score | 0.018935 (rank : 122) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QY61 | Gene names | Irx4, Irxa3 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-4 (Iroquois homeobox protein 4) (Homeodomain protein IRXA3). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.018883 (rank : 123) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
K1C10_HUMAN
|
||||||
| NC score | 0.018462 (rank : 124) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.018362 (rank : 125) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ZFY1_MOUSE
|
||||||
| NC score | 0.018231 (rank : 126) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
RBM14_MOUSE
|
||||||
| NC score | 0.018177 (rank : 127) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
ZFY2_MOUSE
|
||||||
| NC score | 0.018143 (rank : 128) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
PIN1_MOUSE
|
||||||
| NC score | 0.017892 (rank : 129) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
RTN2_HUMAN
|
||||||
| NC score | 0.017822 (rank : 130) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75298, O60509, Q7RTM6, Q7RTN1, Q7RTN2 | Gene names | RTN2, NSPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
RBM19_HUMAN
|
||||||
| NC score | 0.017814 (rank : 131) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.017233 (rank : 132) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.017183 (rank : 133) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.017126 (rank : 134) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.016878 (rank : 135) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.016414 (rank : 136) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
NHRF2_MOUSE
|
||||||
| NC score | 0.016249 (rank : 137) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
TRI55_HUMAN
|
||||||
| NC score | 0.016157 (rank : 138) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYV6, Q53XX3, Q8IUD9, Q8IUE4, Q96DV2, Q96DV3, Q9BYV5 | Gene names | TRIM55, MURF2, RNF29 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 55 (Muscle-specific RING finger protein 2) (MuRF2) (MURF-2) (RING finger protein 29). | |||||
|
ASXL1_MOUSE
|
||||||
| NC score | 0.015940 (rank : 139) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
SDCG1_HUMAN
|
||||||
| NC score | 0.015527 (rank : 140) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
SUHW3_HUMAN
|
||||||
| NC score | 0.015315 (rank : 141) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8ND82, Q9NXR3 | Gene names | SUHW3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.014635 (rank : 142) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
PWP1_MOUSE
|
||||||
| NC score | 0.014551 (rank : 143) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LL5, Q9D6T6 | Gene names | Pwp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periodic tryptophan protein 1 homolog. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.014269 (rank : 144) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.013725 (rank : 145) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.013506 (rank : 146) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
TROP_HUMAN
|
||||||
| NC score | 0.013336 (rank : 147) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.013293 (rank : 148) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
ZN131_HUMAN
|
||||||
| NC score | 0.013171 (rank : 149) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P52739, Q6PIF0 | Gene names | ZNF131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 131. | |||||
|
NECD_HUMAN
|
||||||
| NC score | 0.012504 (rank : 150) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.012499 (rank : 151) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
NKX22_HUMAN
|
||||||
| NC score | 0.012034 (rank : 152) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95096 | Gene names | NKX2-2, NKX2.2, NKX2B | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.2 (Homeobox protein NK-2 homolog B). | |||||
|
PARPT_MOUSE
|
||||||
| NC score | 0.011723 (rank : 153) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.011480 (rank : 154) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.011026 (rank : 155) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.010257 (rank : 156) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
ELMD1_HUMAN
|
||||||
| NC score | 0.010159 (rank : 157) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N336, Q9NPW3 | Gene names | ELMOD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELMO domain-containing protein 1. | |||||
|
41_HUMAN
|
||||||
| NC score | 0.009793 (rank : 158) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
GATA4_HUMAN
|
||||||
| NC score | 0.009737 (rank : 159) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
SYNJ2_MOUSE
|
||||||
| NC score | 0.009675 (rank : 160) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
GNAS1_HUMAN
|
||||||
| NC score | 0.009651 (rank : 161) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.008342 (rank : 162) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
ZN132_HUMAN
|
||||||
| NC score | 0.008238 (rank : 163) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P52740 | Gene names | ZNF132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 132. | |||||
|
FBLN2_HUMAN
|
||||||
| NC score | 0.008238 (rank : 164) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
PHX2B_HUMAN
|
||||||
| NC score | 0.007989 (rank : 165) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99453, Q6PJD9 | Gene names | PHOX2B, PMX2B | |||
|
Domain Architecture |

|
|||||
| Description | Paired mesoderm homeobox protein 2B (Paired-like homeobox 2B) (PHOX2B homeodomain protein) (Neuroblastoma Phox) (NBPhox). | |||||
|
PHX2B_MOUSE
|
||||||
| NC score | 0.007989 (rank : 166) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35690 | Gene names | Phox2b, Pmx2b | |||
|
Domain Architecture |

|
|||||
| Description | Paired mesoderm homeobox protein 2B (Paired-like homeobox 2B) (PHOX2B homeodomain protein) (Neuroblastoma Phox) (NBPhox). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.007279 (rank : 167) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.007230 (rank : 168) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
PEDF_MOUSE
|
||||||
| NC score | 0.006772 (rank : 169) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97298, O70629, O88691 | Gene names | Serpinf1, Pedf, Sdf3 | |||
|
Domain Architecture |

|
|||||
| Description | Pigment epithelium-derived factor precursor (PEDF) (Serpin-F1) (Stromal cell-derived factor 3) (SDF-3) (Caspin). | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.006764 (rank : 170) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SOX3_MOUSE
|
||||||
| NC score | 0.005592 (rank : 171) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
ROBO4_MOUSE
|
||||||
| NC score | 0.005118 (rank : 172) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
HGFA_HUMAN
|
||||||
| NC score | 0.004660 (rank : 173) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
CAMKV_MOUSE
|
||||||
| NC score | 0.003048 (rank : 174) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3UHL1, Q8VD20 | Gene names | Camkv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||