Please be patient as the page loads
|
AKP8L_MOUSE
|
||||||
| SwissProt Accessions | Q9R0L7 | Gene names | Akap8l, Nakap95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AKP8L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985671 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |
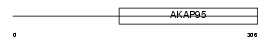
|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
AKP8L_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9R0L7 | Gene names | Akap8l, Nakap95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95). | |||||
|
AKAP8_HUMAN
|
||||||
| θ value | 3.58603e-65 (rank : 3) | NC score | 0.854676 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AKAP8_MOUSE
|
||||||
| θ value | 4.10295e-61 (rank : 4) | NC score | 0.869549 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
ZN326_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 5) | NC score | 0.741762 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 6) | NC score | 0.737275 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 7) | NC score | 0.069955 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 8) | NC score | 0.051052 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.083826 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 10) | NC score | 0.068535 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.089746 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.045864 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.053409 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.059371 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.033484 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.062086 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.041062 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.046186 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
REN3A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.080458 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.070828 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.020719 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ZN516_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.010531 (rank : 99) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.033346 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.038846 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.062207 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.056720 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
K1H2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.023501 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |
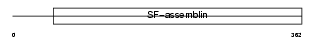
|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.042546 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.039554 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
VDP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.059125 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
TRIM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.026437 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJV3, Q5JYF5, Q8WWK1, Q9UJR9 | Gene names | MID2, FXY2, RNF60, TRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1) (Midin-2) (RING finger protein 60). | |||||
|
TRIM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.026002 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QUS6 | Gene names | Mid2, Fxy2, Trim1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1). | |||||
|
VDP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.060024 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.042759 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.048523 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.062460 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.037570 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RBY1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.034850 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.039315 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.041140 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.022552 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.012520 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.055463 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.042758 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.025993 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
REST_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.033572 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
REST_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.035674 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.035517 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.028367 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.003080 (rank : 107) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.031963 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
GBP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.024028 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01514 | Gene names | Gbp1, Gbp-1, Mag-1, Mpa1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (mGBP1) (mGBP-1) (Interferon-gamma-inducible protein MAG-1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.037843 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.072135 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.032657 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LZTS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.032887 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.025547 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.037689 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.009526 (rank : 100) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.038295 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.036779 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
ASPM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.017972 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.024667 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.020139 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
CE110_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.028661 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
CHM4C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.022813 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D7F7, Q3TVD7, Q9CWH8 | Gene names | Chmp4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 4c (Chromatin-modifying protein 4c) (CHMP4c). | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.020208 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
IP3KA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.018738 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R071 | Gene names | Itpka | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase A (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase A) (IP3K A) (IP3 3-kinase A). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.014636 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.019748 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.042279 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.024976 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
RBBP5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.012274 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |
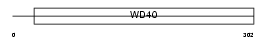
|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
ZN335_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.004171 (rank : 106) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.047921 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
ZSCA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | -0.000445 (rank : 108) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07230 | Gene names | Zscan2, Zfp-29, Zfp29 | |||
|
Domain Architecture |
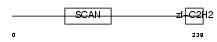
|
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29) (Zfp-29). | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.034907 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.011221 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
FETA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.014628 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02771 | Gene names | AFP | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein precursor (Alpha-fetoglobulin) (Alpha-1- fetoprotein). | |||||
|
IP3KA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.017457 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23677, Q8TAN3 | Gene names | ITPKA | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase A (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase A) (IP3K A) (IP3 3-kinase A). | |||||
|
LRFN5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.005267 (rank : 104) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXA0, Q8BJH4, Q8BZL0 | Gene names | Lrfn5 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 5 precursor. | |||||
|
NCOA5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.023793 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91W39 | Gene names | Ncoa5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.007903 (rank : 101) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.014119 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
RRMJ3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.021137 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.010971 (rank : 98) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
UBP2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.027176 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |
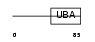
|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
UBP2L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.027823 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80X50, Q8BIT6, Q8BIW4, Q8BJ01, Q8CIG7, Q8K102, Q9CRT6 | Gene names | Ubap2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 2-like. | |||||
|
ZN462_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.004629 (rank : 105) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96JM2, Q8N408 | Gene names | ZNF462, KIAA1803 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 462. | |||||
|
ARRD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.007727 (rank : 102) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TBH0, O95895, Q6ZRV9, Q8WYG6 | Gene names | ARRDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 2. | |||||
|
HD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.015634 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
K1H6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.014657 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O76013 | Gene names | KRTHA6, HHA6, HKA6 | |||
|
Domain Architecture |
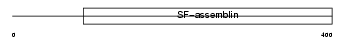
|
|||||
| Description | Keratin, type I cuticular Ha6 (Hair keratin, type I Ha6). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.013392 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.023296 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.006665 (rank : 103) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.062720 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.054311 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
LORI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.058441 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.056934 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.055784 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.064838 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.070940 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.051696 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.053330 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.053321 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.052545 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.050524 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.050004 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
AKP8L_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9R0L7 | Gene names | Akap8l, Nakap95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95). | |||||
|
AKP8L_HUMAN
|
||||||
| NC score | 0.985671 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
AKAP8_MOUSE
|
||||||
| NC score | 0.869549 (rank : 3) | θ value | 4.10295e-61 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AKAP8_HUMAN
|
||||||
| NC score | 0.854676 (rank : 4) | θ value | 3.58603e-65 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
ZN326_HUMAN
|
||||||
| NC score | 0.741762 (rank : 5) | θ value | 4.42448e-31 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.737275 (rank : 6) | θ value | 3.74554e-30 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.089746 (rank : 7) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.083826 (rank : 8) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
REN3A_HUMAN
|
||||||
| NC score | 0.080458 (rank : 9) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.072135 (rank : 10) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.070940 (rank : 11) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.070828 (rank : 12) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.069955 (rank : 13) | θ value | 0.0148317 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.068535 (rank : 14) | θ value | 0.0330416 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.064838 (rank : 15) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.062720 (rank : 16) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.062460 (rank : 17) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.062207 (rank : 18) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.062086 (rank : 19) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
VDP_HUMAN
|
||||||
| NC score | 0.060024 (rank : 20) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.059371 (rank : 21) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
VDP_MOUSE
|
||||||
| NC score | 0.059125 (rank : 22) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.058441 (rank : 23) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.056934 (rank : 24) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.056720 (rank : 25) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.055784 (rank : 26) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.055463 (rank : 27) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.054311 (rank : 28) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.053409 (rank : 29) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.053330 (rank : 30) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.053321 (rank : 31) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.052545 (rank : 32) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.051696 (rank : 33) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.051052 (rank : 34) | θ value | 0.0252991 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.050524 (rank : 35) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.050004 (rank : 36) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.048523 (rank : 37) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.047921 (rank : 38) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.046186 (rank : 39) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.045864 (rank : 40) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.042759 (rank : 41) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.042758 (rank : 42) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.042546 (rank : 43) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.042279 (rank : 44) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.041140 (rank : 45) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.041062 (rank : 46) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.039554 (rank : 47) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.039315 (rank : 48) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.038846 (rank : 49) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.038295 (rank : 50) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.037843 (rank : 51) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.037689 (rank : 52) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.037570 (rank : 53) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.036779 (rank : 54) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.035674 (rank : 55) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.035517 (rank : 56) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.034907 (rank : 57) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
RBY1A_HUMAN
|
||||||
| NC score | 0.034850 (rank : 58) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.033572 (rank : 59) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.033484 (rank : 60) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.033346 (rank : 61) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
LZTS2_MOUSE
|
||||||
| NC score | 0.032887 (rank : 62) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.032657 (rank : 63) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.031963 (rank : 64) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
CE110_HUMAN
|
||||||
| NC score | 0.028661 (rank : 65) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.028367 (rank : 66) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
UBP2L_MOUSE
|
||||||
| NC score | 0.027823 (rank : 67) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80X50, Q8BIT6, Q8BIW4, Q8BJ01, Q8CIG7, Q8K102, Q9CRT6 | Gene names | Ubap2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 2-like. | |||||
|
UBP2L_HUMAN
|
||||||
| NC score | 0.027176 (rank : 68) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
TRIM1_HUMAN
|
||||||
| NC score | 0.026437 (rank : 69) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJV3, Q5JYF5, Q8WWK1, Q9UJR9 | Gene names | MID2, FXY2, RNF60, TRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1) (Midin-2) (RING finger protein 60). | |||||
|
TRIM1_MOUSE
|
||||||
| NC score | 0.026002 (rank : 70) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QUS6 | Gene names | Mid2, Fxy2, Trim1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.025993 (rank : 71) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.025547 (rank : 72) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.024976 (rank : 73) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.024667 (rank : 74) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
GBP1_MOUSE
|
||||||
| NC score | 0.024028 (rank : 75) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01514 | Gene names | Gbp1, Gbp-1, Mag-1, Mpa1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (mGBP1) (mGBP-1) (Interferon-gamma-inducible protein MAG-1). | |||||
|
NCOA5_MOUSE
|
||||||
| NC score | 0.023793 (rank : 76) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91W39 | Gene names | Ncoa5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
K1H2_MOUSE
|
||||||
| NC score | 0.023501 (rank : 77) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.023296 (rank : 78) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CHM4C_MOUSE
|
||||||
| NC score | 0.022813 (rank : 79) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D7F7, Q3TVD7, Q9CWH8 | Gene names | Chmp4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 4c (Chromatin-modifying protein 4c) (CHMP4c). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.022552 (rank : 80) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
RRMJ3_MOUSE
|
||||||
| NC score | 0.021137 (rank : 81) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.020719 (rank : 82) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.020208 (rank : 83) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.020139 (rank : 84) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.019748 (rank : 85) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
IP3KA_MOUSE
|
||||||
| NC score | 0.018738 (rank : 86) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R071 | Gene names | Itpka | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase A (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase A) (IP3K A) (IP3 3-kinase A). | |||||
|
ASPM_HUMAN
|
||||||
| NC score | 0.017972 (rank : 87) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||
|
IP3KA_HUMAN
|
||||||
| NC score | 0.017457 (rank : 88) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23677, Q8TAN3 | Gene names | ITPKA | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase A (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase A) (IP3K A) (IP3 3-kinase A). | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.015634 (rank : 89) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
K1H6_HUMAN
|
||||||
| NC score | 0.014657 (rank : 90) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O76013 | Gene names | KRTHA6, HHA6, HKA6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha6 (Hair keratin, type I Ha6). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.014636 (rank : 91) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
FETA_HUMAN
|
||||||
| NC score | 0.014628 (rank : 92) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02771 | Gene names | AFP | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein precursor (Alpha-fetoglobulin) (Alpha-1- fetoprotein). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.014119 (rank : 93) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.013392 (rank : 94) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.012520 (rank : 95) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
RBBP5_HUMAN
|
||||||
| NC score | 0.012274 (rank : 96) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.011221 (rank : 97) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.010971 (rank : 98) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
ZN516_HUMAN
|
||||||
| NC score | 0.010531 (rank : 99) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.009526 (rank : 100) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.007903 (rank : 101) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ARRD2_HUMAN
|
||||||
| NC score | 0.007727 (rank : 102) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TBH0, O95895, Q6ZRV9, Q8WYG6 | Gene names | ARRDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 2. | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.006665 (rank : 103) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
LRFN5_MOUSE
|
||||||
| NC score | 0.005267 (rank : 104) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXA0, Q8BJH4, Q8BZL0 | Gene names | Lrfn5 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 5 precursor. | |||||
|
ZN462_HUMAN
|
||||||
| NC score | 0.004629 (rank : 105) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96JM2, Q8N408 | Gene names | ZNF462, KIAA1803 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 462. | |||||
|
ZN335_HUMAN
|
||||||
| NC score | 0.004171 (rank : 106) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.003080 (rank : 107) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
ZSCA2_MOUSE
|
||||||
| NC score | -0.000445 (rank : 108) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07230 | Gene names | Zscan2, Zfp-29, Zfp29 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29) (Zfp-29). | |||||