Please be patient as the page loads
|
AKP8L_HUMAN
|
||||||
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AKP8L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
AKP8L_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.985671 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R0L7 | Gene names | Akap8l, Nakap95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95). | |||||
|
AKAP8_HUMAN
|
||||||
| θ value | 1.15359e-63 (rank : 3) | NC score | 0.857550 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AKAP8_MOUSE
|
||||||
| θ value | 2.03627e-60 (rank : 4) | NC score | 0.875568 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
ZN326_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 5) | NC score | 0.743217 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 6) | NC score | 0.738700 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 7) | NC score | 0.060356 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.077992 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.082714 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.057731 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.060661 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.039907 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.043266 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.044087 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.020932 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
GP176_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.015856 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 522 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14439, Q6NXF6 | Gene names | GPR176 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (HB-954). | |||||
|
TRIM1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.029617 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJV3, Q5JYF5, Q8WWK1, Q9UJR9 | Gene names | MID2, FXY2, RNF60, TRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1) (Midin-2) (RING finger protein 60). | |||||
|
TRIM1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.029129 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QUS6 | Gene names | Mid2, Fxy2, Trim1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.060156 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
K1H2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.016895 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
VDP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.051515 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
ABCA4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.010478 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35600 | Gene names | Abca4, Abcr | |||
|
Domain Architecture |

|
|||||
| Description | Retinal-specific ATP-binding cassette transporter (ATP-binding cassette sub-family A member 4) (RIM ABC transporter) (RIM protein) (RmP). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.030522 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
SMG5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.034487 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
VDP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.048665 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.034786 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.059854 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.069782 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
ZN516_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.009963 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
ZN598_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.026684 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80YR4, Q6KAT0, Q8R3S1 | Gene names | Znf598, Zfp598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.019022 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CPSF2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.032192 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2I0, Q6NSJ1, Q9H3W7 | Gene names | CPSF2, CPSF100, KIAA1367 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 100 kDa subunit (CPSF 100 kDa subunit). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.030600 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
RBY1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.035348 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
ZN335_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.005998 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
GBP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.021135 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01514 | Gene names | Gbp1, Gbp-1, Mag-1, Mpa1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (mGBP1) (mGBP-1) (Interferon-gamma-inducible protein MAG-1). | |||||
|
LRFN5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.006936 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXA0, Q8BJH4, Q8BZL0 | Gene names | Lrfn5 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 5 precursor. | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.040783 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
RBP56_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.058029 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.040519 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.009157 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
CASC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.032002 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15234 | Gene names | CASC3, MLN51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein) (Metastatic lymph node protein 51) (Protein MLN 51) (Protein barentsz) (Btz). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.024388 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GP176_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.012779 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80WT4, Q80UC4 | Gene names | Gpr176, Agr9, Gm1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (G-protein coupled receptor AGR9). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.012666 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
ASPM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.017618 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||
|
CHM4C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.022967 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7F7, Q3TVD7, Q9CWH8 | Gene names | Chmp4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 4c (Chromatin-modifying protein 4c) (CHMP4c). | |||||
|
CPSF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.028885 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35218 | Gene names | Cpsf2, Cpsf100, Mcpsf | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 100 kDa subunit (CPSF 100 kDa subunit). | |||||
|
IEX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.024442 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P46695, Q92691, Q93044 | Gene names | IER3, DIF2, IEX1, PRG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Radiation-inducible immediate-early gene IEX-1 (Immediate early protein GLY96) (Immediate early response 3 protein) (PACAP-responsive gene 1 protein) (Protein PRG1) (Differentiation-dependent gene 2 protein) (Protein DIF-2). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.047338 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.019365 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.022701 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.007898 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.034640 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.011251 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.035285 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.008256 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.039122 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ARP8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.018102 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H981, Q9H663 | Gene names | ACTR8 | |||
|
Domain Architecture |

|
|||||
| Description | Actin-related protein 8. | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.021234 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.021731 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
CE110_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.024908 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.010121 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.022236 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
FETA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.014832 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02771 | Gene names | AFP | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein precursor (Alpha-fetoglobulin) (Alpha-1- fetoprotein). | |||||
|
HA1T_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.001250 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14432, Q31194, Q31203 | Gene names | H2-T3 | |||
|
Domain Architecture |
|
|||||
| Description | H-2 class I histocompatibility antigen, TLA(B) alpha chain precursor (MHC thymus leukemia antigen). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.045640 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
LY10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.012282 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.019251 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
NCOA5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.023626 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91W39 | Gene names | Ncoa5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.011885 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ROA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.027049 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BG05, Q8BHF8 | Gene names | Hnrpa3, Hnrnpa3 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
RX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.003501 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35602, O08748 | Gene names | Rax, Rx | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.007329 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.041057 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
ARAF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | -0.001260 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||
|
ARP8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.017013 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2S9, Q9CYC4 | Gene names | Actr8 | |||
|
Domain Architecture |

|
|||||
| Description | Actin-related protein 8. | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.005479 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.017026 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
LIMD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.004931 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.039642 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
NFM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.028784 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.016727 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.022535 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.007180 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.027975 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ZFY1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.003038 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
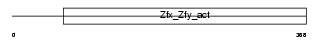
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
ZFY2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.003148 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
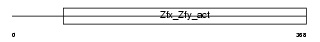
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
ZN462_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.005511 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JM2, Q8N408 | Gene names | ZNF462, KIAA1803 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 462. | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.060141 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.050567 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.062646 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
FUS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.054564 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
LORI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.062347 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.058939 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
REN3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.055050 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
AKP8L_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
AKP8L_MOUSE
|
||||||
| NC score | 0.985671 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R0L7 | Gene names | Akap8l, Nakap95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95). | |||||
|
AKAP8_MOUSE
|
||||||
| NC score | 0.875568 (rank : 3) | θ value | 2.03627e-60 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AKAP8_HUMAN
|
||||||
| NC score | 0.857550 (rank : 4) | θ value | 1.15359e-63 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
ZN326_HUMAN
|
||||||
| NC score | 0.743217 (rank : 5) | θ value | 1.08979e-29 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.738700 (rank : 6) | θ value | 3.50572e-28 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.082714 (rank : 7) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.077992 (rank : 8) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.069782 (rank : 9) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.062646 (rank : 10) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.062347 (rank : 11) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.060661 (rank : 12) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.060356 (rank : 13) | θ value | 0.00134147 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.060156 (rank : 14) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.060141 (rank : 15) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.059854 (rank : 16) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.058939 (rank : 17) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
RBP56_HUMAN
|
||||||
| NC score | 0.058029 (rank : 18) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.057731 (rank : 19) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
REN3A_HUMAN
|
||||||
| NC score | 0.055050 (rank : 20) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.054564 (rank : 21) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
VDP_MOUSE
|
||||||
| NC score | 0.051515 (rank : 22) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.050567 (rank : 23) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
VDP_HUMAN
|
||||||
| NC score | 0.048665 (rank : 24) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.047338 (rank : 25) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.045640 (rank : 26) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.044087 (rank : 27) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.043266 (rank : 28) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.041057 (rank : 29) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.040783 (rank : 30) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.040519 (rank : 31) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.039907 (rank : 32) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.039642 (rank : 33) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.039122 (rank : 34) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
RBY1A_HUMAN
|
||||||
| NC score | 0.035348 (rank : 35) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.035285 (rank : 36) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.034786 (rank : 37) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.034640 (rank : 38) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
SMG5_HUMAN
|
||||||
| NC score | 0.034487 (rank : 39) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
CPSF2_HUMAN
|
||||||
| NC score | 0.032192 (rank : 40) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2I0, Q6NSJ1, Q9H3W7 | Gene names | CPSF2, CPSF100, KIAA1367 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 100 kDa subunit (CPSF 100 kDa subunit). | |||||
|
CASC3_HUMAN
|
||||||
| NC score | 0.032002 (rank : 41) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15234 | Gene names | CASC3, MLN51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein) (Metastatic lymph node protein 51) (Protein MLN 51) (Protein barentsz) (Btz). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.030600 (rank : 42) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.030522 (rank : 43) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
TRIM1_HUMAN
|
||||||
| NC score | 0.029617 (rank : 44) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJV3, Q5JYF5, Q8WWK1, Q9UJR9 | Gene names | MID2, FXY2, RNF60, TRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1) (Midin-2) (RING finger protein 60). | |||||
|
TRIM1_MOUSE
|
||||||
| NC score | 0.029129 (rank : 45) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QUS6 | Gene names | Mid2, Fxy2, Trim1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1). | |||||
|
CPSF2_MOUSE
|
||||||
| NC score | 0.028885 (rank : 46) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35218 | Gene names | Cpsf2, Cpsf100, Mcpsf | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 100 kDa subunit (CPSF 100 kDa subunit). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.028784 (rank : 47) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.027975 (rank : 48) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ROA3_MOUSE
|
||||||
| NC score | 0.027049 (rank : 49) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BG05, Q8BHF8 | Gene names | Hnrpa3, Hnrnpa3 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
ZN598_MOUSE
|
||||||
| NC score | 0.026684 (rank : 50) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80YR4, Q6KAT0, Q8R3S1 | Gene names | Znf598, Zfp598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
CE110_HUMAN
|
||||||
| NC score | 0.024908 (rank : 51) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
IEX1_HUMAN
|
||||||
| NC score | 0.024442 (rank : 52) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P46695, Q92691, Q93044 | Gene names | IER3, DIF2, IEX1, PRG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Radiation-inducible immediate-early gene IEX-1 (Immediate early protein GLY96) (Immediate early response 3 protein) (PACAP-responsive gene 1 protein) (Protein PRG1) (Differentiation-dependent gene 2 protein) (Protein DIF-2). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.024388 (rank : 53) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
NCOA5_MOUSE
|
||||||
| NC score | 0.023626 (rank : 54) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91W39 | Gene names | Ncoa5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
CHM4C_MOUSE
|
||||||
| NC score | 0.022967 (rank : 55) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7F7, Q3TVD7, Q9CWH8 | Gene names | Chmp4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 4c (Chromatin-modifying protein 4c) (CHMP4c). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.022701 (rank : 56) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.022535 (rank : 57) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.022236 (rank : 58) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.021731 (rank : 59) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.021234 (rank : 60) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
GBP1_MOUSE
|
||||||
| NC score | 0.021135 (rank : 61) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01514 | Gene names | Gbp1, Gbp-1, Mag-1, Mpa1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (mGBP1) (mGBP-1) (Interferon-gamma-inducible protein MAG-1). | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.020932 (rank : 62) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.019365 (rank : 63) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.019251 (rank : 64) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.019022 (rank : 65) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ARP8_HUMAN
|
||||||
| NC score | 0.018102 (rank : 66) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H981, Q9H663 | Gene names | ACTR8 | |||
|
Domain Architecture |

|
|||||
| Description | Actin-related protein 8. | |||||
|
ASPM_HUMAN
|
||||||
| NC score | 0.017618 (rank : 67) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.017026 (rank : 68) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
ARP8_MOUSE
|
||||||
| NC score | 0.017013 (rank : 69) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2S9, Q9CYC4 | Gene names | Actr8 | |||
|
Domain Architecture |

|
|||||
| Description | Actin-related protein 8. | |||||
|
K1H2_MOUSE
|
||||||
| NC score | 0.016895 (rank : 70) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.016727 (rank : 71) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
GP176_HUMAN
|
||||||
| NC score | 0.015856 (rank : 72) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 522 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14439, Q6NXF6 | Gene names | GPR176 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (HB-954). | |||||
|
FETA_HUMAN
|
||||||
| NC score | 0.014832 (rank : 73) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02771 | Gene names | AFP | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein precursor (Alpha-fetoglobulin) (Alpha-1- fetoprotein). | |||||
|
GP176_MOUSE
|
||||||
| NC score | 0.012779 (rank : 74) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80WT4, Q80UC4 | Gene names | Gpr176, Agr9, Gm1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (G-protein coupled receptor AGR9). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.012666 (rank : 75) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.012282 (rank : 76) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.011885 (rank : 77) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.011251 (rank : 78) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
ABCA4_MOUSE
|
||||||
| NC score | 0.010478 (rank : 79) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35600 | Gene names | Abca4, Abcr | |||
|
Domain Architecture |

|
|||||
| Description | Retinal-specific ATP-binding cassette transporter (ATP-binding cassette sub-family A member 4) (RIM ABC transporter) (RIM protein) (RmP). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.010121 (rank : 80) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
ZN516_HUMAN
|
||||||
| NC score | 0.009963 (rank : 81) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.009157 (rank : 82) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.008256 (rank : 83) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.007898 (rank : 84) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.007329 (rank : 85) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.007180 (rank : 86) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
LRFN5_MOUSE
|
||||||
| NC score | 0.006936 (rank : 87) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXA0, Q8BJH4, Q8BZL0 | Gene names | Lrfn5 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 5 precursor. | |||||
|
ZN335_HUMAN
|
||||||
| NC score | 0.005998 (rank : 88) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
ZN462_HUMAN
|
||||||
| NC score | 0.005511 (rank : 89) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JM2, Q8N408 | Gene names | ZNF462, KIAA1803 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 462. | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.005479 (rank : 90) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
LIMD1_HUMAN
|
||||||
| NC score | 0.004931 (rank : 91) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
RX_MOUSE
|
||||||
| NC score | 0.003501 (rank : 92) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35602, O08748 | Gene names | Rax, Rx | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
ZFY2_MOUSE
|
||||||
| NC score | 0.003148 (rank : 93) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
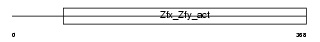
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
ZFY1_MOUSE
|
||||||
| NC score | 0.003038 (rank : 94) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
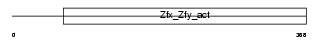
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
HA1T_MOUSE
|
||||||
| NC score | 0.001250 (rank : 95) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14432, Q31194, Q31203 | Gene names | H2-T3 | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class I histocompatibility antigen, TLA(B) alpha chain precursor (MHC thymus leukemia antigen). | |||||
|
ARAF_HUMAN
|
||||||
| NC score | -0.001260 (rank : 96) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||