Please be patient as the page loads
|
CHM4C_MOUSE
|
||||||
| SwissProt Accessions | Q9D7F7, Q3TVD7, Q9CWH8 | Gene names | Chmp4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 4c (Chromatin-modifying protein 4c) (CHMP4c). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CHM4C_MOUSE
|
||||||
| θ value | 6.95003e-85 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9D7F7, Q3TVD7, Q9CWH8 | Gene names | Chmp4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 4c (Chromatin-modifying protein 4c) (CHMP4c). | |||||
|
CHM4C_HUMAN
|
||||||
| θ value | 4.22625e-74 (rank : 2) | NC score | 0.986388 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96CF2 | Gene names | CHMP4C, SHAX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 4c (Chromatin-modifying protein 4c) (CHMP4c) (Vacuolar protein sorting 7-3) (SNF7-3) (hSnf7-3) (SNF7 homolog associated with Alix 3). | |||||
|
CHM4B_HUMAN
|
||||||
| θ value | 3.37202e-47 (rank : 3) | NC score | 0.953256 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H444, Q53ZD6 | Gene names | CHMP4B, C20orf178, SHAX1 | |||
|
Domain Architecture |
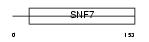
|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b) (Vacuolar protein sorting 7-2) (SNF7-2) (hSnf7-2) (SNF7 homolog associated with Alix 1) (hVps32). | |||||
|
CHM4B_MOUSE
|
||||||
| θ value | 3.37202e-47 (rank : 4) | NC score | 0.953007 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D8B3, Q3TXM7, Q91VM7, Q922P1 | Gene names | Chmp4b | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b). | |||||
|
CG301_HUMAN
|
||||||
| θ value | 1.46607e-42 (rank : 5) | NC score | 0.933646 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59074, Q8NFA9 | Gene names | ames=CGI-301 | |||
|
Domain Architecture |
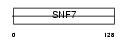
|
|||||
| Description | Protein CGI-301. | |||||
|
CHM4A_HUMAN
|
||||||
| θ value | 5.39604e-37 (rank : 6) | NC score | 0.920473 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BY43, Q32Q79, Q86SZ8, Q96QJ9, Q9P026 | Gene names | CHMP4A, C14orf123, SHAX2 | |||
|
Domain Architecture |
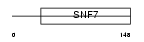
|
|||||
| Description | Charged multivesicular body protein 4a (Chromatin-modifying protein 4a) (CHMP4a) (Vacuolar protein sorting 7-1) (SNF7-1) (hSnf-1) (SNF7 homolog associated with Alix-2). | |||||
|
CHMP7_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 7) | NC score | 0.496594 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WUX9, Q8NDM1, Q9BT50 | Gene names | CHMP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CHMP7. | |||||
|
CHMP7_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 8) | NC score | 0.473265 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R1T1, Q8CFW4 | Gene names | Chmp7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CHMP7. | |||||
|
CHMP6_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 9) | NC score | 0.431677 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
CHMP6_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 10) | NC score | 0.331826 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96FZ7, Q53FU4, Q9HAE8 | Gene names | CHMP6, VPS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6) (Vacuolar protein sorting-associated protein 20) (hVps20). | |||||
|
BICD2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 11) | NC score | 0.080712 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.081227 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CNGA2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.034770 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 14) | NC score | 0.059773 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.067176 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.070495 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.064716 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.082638 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
CF152_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.065775 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.061093 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.045356 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.031397 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.054442 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
KIF1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.040327 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12756 | Gene names | KIF1A, ATSV | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
KIF1A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.040271 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P33173, Q61770 | Gene names | Kif1a, Atsv, Kif1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.051978 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
SDPR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.065895 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.053581 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.051736 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ANGP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.029352 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15123, Q9NRR7, Q9P2Y7 | Gene names | ANGPT2 | |||
|
Domain Architecture |
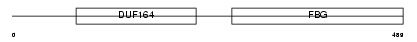
|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.063479 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.052388 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.058812 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.030049 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
STIM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.053911 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.047263 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
TAOK3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.017621 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1494 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H2K8, Q658N1, Q8IUM4, Q9HC79, Q9NZM9, Q9UHG7 | Gene names | TAOK3, DPK, JIK, KDS, MAP3K18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3) (Jun kinase-inhibitory kinase) (JNK/SAPK- inhibitory kinase) (Dendritic cell-derived protein kinase) (Cutaneous T-cell lymphoma tumor antigen HD-CL-09) (CTCL tumor antigen HD-CL-09) (Kinase from chicken homolog A) (hKFC-A). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.041424 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.049499 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CCD11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.062665 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CHMP5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.251188 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZZ3, Q5VXW2, Q96AV2, Q9HB68, Q9NYS4, Q9Y323 | Gene names | CHMP5, C9orf83, SNF7DC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 5 (Chromatin-modifying protein 5) (Vacuolar protein sorting 60) (Vps60) (hVps60) (SNF7 domain-containing protein 2). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.028205 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.075988 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.042103 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.042275 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
STIM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.052285 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
TMCO3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.048827 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BH01, Q3UFX1, Q7TMT5, Q8BGL4, Q8R303 | Gene names | Tmco3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 3 precursor. | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.053619 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
IFT88_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.049342 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
LRSM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.042700 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80ZI6 | Gene names | Lrsam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase). | |||||
|
AKP8L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.022967 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |
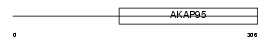
|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
AKP8L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.022813 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0L7 | Gene names | Akap8l, Nakap95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.072106 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
KIF1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.033509 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60333, Q96Q94, Q9BV80, Q9P280 | Gene names | KIF1B, KIAA0591, KIAA1448 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B (Klp). | |||||
|
KIF1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.033508 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60575, Q9R0B4, Q9WVE5, Q9Z119 | Gene names | Kif1b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B. | |||||
|
EMID2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.007143 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VF6, Q8K4P3 | Gene names | Emid2, Col26a, Col26a1, Emu2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
IFT88_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.041861 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
LRMP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.022810 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12912, Q8N301 | Gene names | LRMP, JAW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
TMCO3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.039869 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6UWJ1, Q5JSB1, Q6NUN1, Q8NG29, Q8TCI6, Q96EA6, Q9NWT2 | Gene names | TMCO3, C13orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 3 precursor (Putative LAG1-interacting protein). | |||||
|
ANGP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.021314 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
HCDH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.010668 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
Domain Architecture |
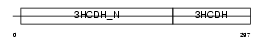
|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.036445 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
TRAF3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.019581 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60803, Q62380 | Gene names | Traf3, Craf1, Trafamn | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 3 (CD40 receptor-associated factor 1) (CRAF1) (TRAFAMN). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.038123 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TRI13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.005089 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CYB0, Q8CEV0, Q923J0, Q925P1, Q99PQ0 | Gene names | Trim13, Rfp2 | |||
|
Domain Architecture |
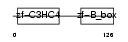
|
|||||
| Description | Tripartite motif-containing protein 13 (Ret finger protein 2) (Putative tumor suppressor RFP2). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.060857 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.060755 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CALD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.051280 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CCD11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.052542 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D439 | Gene names | Ccdc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CF152_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.052363 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
CHMP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.202843 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D7S9, Q3UI64 | Gene names | Chmp5, Snf7dc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 5 (Chromatin-modifying protein 5) (SNF7 domain-containing protein 2). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.050566 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.059074 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.052158 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.053137 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.050578 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
INCE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.050094 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.052797 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
NDE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.067108 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.050050 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CHM4C_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.95003e-85 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9D7F7, Q3TVD7, Q9CWH8 | Gene names | Chmp4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 4c (Chromatin-modifying protein 4c) (CHMP4c). | |||||
|
CHM4C_HUMAN
|
||||||
| NC score | 0.986388 (rank : 2) | θ value | 4.22625e-74 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96CF2 | Gene names | CHMP4C, SHAX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 4c (Chromatin-modifying protein 4c) (CHMP4c) (Vacuolar protein sorting 7-3) (SNF7-3) (hSnf7-3) (SNF7 homolog associated with Alix 3). | |||||
|
CHM4B_HUMAN
|
||||||
| NC score | 0.953256 (rank : 3) | θ value | 3.37202e-47 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H444, Q53ZD6 | Gene names | CHMP4B, C20orf178, SHAX1 | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b) (Vacuolar protein sorting 7-2) (SNF7-2) (hSnf7-2) (SNF7 homolog associated with Alix 1) (hVps32). | |||||
|
CHM4B_MOUSE
|
||||||
| NC score | 0.953007 (rank : 4) | θ value | 3.37202e-47 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D8B3, Q3TXM7, Q91VM7, Q922P1 | Gene names | Chmp4b | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b). | |||||
|
CG301_HUMAN
|
||||||
| NC score | 0.933646 (rank : 5) | θ value | 1.46607e-42 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59074, Q8NFA9 | Gene names | ames=CGI-301 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CGI-301. | |||||
|
CHM4A_HUMAN
|
||||||
| NC score | 0.920473 (rank : 6) | θ value | 5.39604e-37 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BY43, Q32Q79, Q86SZ8, Q96QJ9, Q9P026 | Gene names | CHMP4A, C14orf123, SHAX2 | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4a (Chromatin-modifying protein 4a) (CHMP4a) (Vacuolar protein sorting 7-1) (SNF7-1) (hSnf-1) (SNF7 homolog associated with Alix-2). | |||||
|
CHMP7_HUMAN
|
||||||
| NC score | 0.496594 (rank : 7) | θ value | 3.08544e-08 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WUX9, Q8NDM1, Q9BT50 | Gene names | CHMP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CHMP7. | |||||
|
CHMP7_MOUSE
|
||||||
| NC score | 0.473265 (rank : 8) | θ value | 2.88788e-06 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R1T1, Q8CFW4 | Gene names | Chmp7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CHMP7. | |||||
|
CHMP6_MOUSE
|
||||||
| NC score | 0.431677 (rank : 9) | θ value | 0.00175202 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
CHMP6_HUMAN
|
||||||
| NC score | 0.331826 (rank : 10) | θ value | 0.00665767 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96FZ7, Q53FU4, Q9HAE8 | Gene names | CHMP6, VPS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6) (Vacuolar protein sorting-associated protein 20) (hVps20). | |||||
|
CHMP5_HUMAN
|
||||||
| NC score | 0.251188 (rank : 11) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZZ3, Q5VXW2, Q96AV2, Q9HB68, Q9NYS4, Q9Y323 | Gene names | CHMP5, C9orf83, SNF7DC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 5 (Chromatin-modifying protein 5) (Vacuolar protein sorting 60) (Vps60) (hVps60) (SNF7 domain-containing protein 2). | |||||
|
CHMP5_MOUSE
|
||||||
| NC score | 0.202843 (rank : 12) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D7S9, Q3UI64 | Gene names | Chmp5, Snf7dc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 5 (Chromatin-modifying protein 5) (SNF7 domain-containing protein 2). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.082638 (rank : 13) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.081227 (rank : 14) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
BICD2_HUMAN
|
||||||
| NC score | 0.080712 (rank : 15) | θ value | 0.0252991 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.075988 (rank : 16) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.072106 (rank : 17) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.070495 (rank : 18) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.067176 (rank : 19) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
NDE1_HUMAN
|
||||||
| NC score | 0.067108 (rank : 20) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
SDPR_HUMAN
|
||||||
| NC score | 0.065895 (rank : 21) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.065775 (rank : 22) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.064716 (rank : 23) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.063479 (rank : 24) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.062665 (rank : 25) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.061093 (rank : 26) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.060857 (rank : 27) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.060755 (rank : 28) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.059773 (rank : 29) | θ value | 0.0736092 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.059074 (rank : 30) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.058812 (rank : 31) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.054442 (rank : 32) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
STIM1_HUMAN
|
||||||
| NC score | 0.053911 (rank : 33) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.053619 (rank : 34) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.053581 (rank : 35) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.053137 (rank : 36) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.052797 (rank : 37) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
CCD11_MOUSE
|
||||||
| NC score | 0.052542 (rank : 38) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D439 | Gene names | Ccdc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.052388 (rank : 39) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CF152_MOUSE
|
||||||
| NC score | 0.052363 (rank : 40) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
STIM1_MOUSE
|
||||||
| NC score | 0.052285 (rank : 41) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.052158 (rank : 42) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.051978 (rank : 43) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.051736 (rank : 44) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.051280 (rank : 45) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.050578 (rank : 46) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.050566 (rank : 47) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.050094 (rank : 48) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.050050 (rank : 49) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.049499 (rank : 50) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
IFT88_HUMAN
|
||||||
| NC score | 0.049342 (rank : 51) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
TMCO3_MOUSE
|
||||||
| NC score | 0.048827 (rank : 52) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BH01, Q3UFX1, Q7TMT5, Q8BGL4, Q8R303 | Gene names | Tmco3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 3 precursor. | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.047263 (rank : 53) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.045356 (rank : 54) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
LRSM1_MOUSE
|
||||||
| NC score | 0.042700 (rank : 55) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80ZI6 | Gene names | Lrsam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.042275 (rank : 56) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.042103 (rank : 57) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
IFT88_MOUSE
|
||||||
| NC score | 0.041861 (rank : 58) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.041424 (rank : 59) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
KIF1A_HUMAN
|
||||||
| NC score | 0.040327 (rank : 60) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12756 | Gene names | KIF1A, ATSV | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
KIF1A_MOUSE
|
||||||
| NC score | 0.040271 (rank : 61) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P33173, Q61770 | Gene names | Kif1a, Atsv, Kif1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
TMCO3_HUMAN
|
||||||
| NC score | 0.039869 (rank : 62) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6UWJ1, Q5JSB1, Q6NUN1, Q8NG29, Q8TCI6, Q96EA6, Q9NWT2 | Gene names | TMCO3, C13orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 3 precursor (Putative LAG1-interacting protein). | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.038123 (rank : 63) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.036445 (rank : 64) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
CNGA2_MOUSE
|
||||||
| NC score | 0.034770 (rank : 65) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
KIF1B_HUMAN
|
||||||
| NC score | 0.033509 (rank : 66) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60333, Q96Q94, Q9BV80, Q9P280 | Gene names | KIF1B, KIAA0591, KIAA1448 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B (Klp). | |||||
|
KIF1B_MOUSE
|
||||||
| NC score | 0.033508 (rank : 67) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60575, Q9R0B4, Q9WVE5, Q9Z119 | Gene names | Kif1b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B. | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.031397 (rank : 68) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.030049 (rank : 69) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
ANGP2_HUMAN
|
||||||
| NC score | 0.029352 (rank : 70) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15123, Q9NRR7, Q9P2Y7 | Gene names | ANGPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.028205 (rank : 71) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
AKP8L_HUMAN
|
||||||
| NC score | 0.022967 (rank : 72) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
AKP8L_MOUSE
|
||||||
| NC score | 0.022813 (rank : 73) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0L7 | Gene names | Akap8l, Nakap95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95). | |||||
|
LRMP_HUMAN
|
||||||
| NC score | 0.022810 (rank : 74) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12912, Q8N301 | Gene names | LRMP, JAW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
ANGP2_MOUSE
|
||||||
| NC score | 0.021314 (rank : 75) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
TRAF3_MOUSE
|
||||||
| NC score | 0.019581 (rank : 76) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60803, Q62380 | Gene names | Traf3, Craf1, Trafamn | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 3 (CD40 receptor-associated factor 1) (CRAF1) (TRAFAMN). | |||||
|
TAOK3_HUMAN
|
||||||
| NC score | 0.017621 (rank : 77) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1494 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H2K8, Q658N1, Q8IUM4, Q9HC79, Q9NZM9, Q9UHG7 | Gene names | TAOK3, DPK, JIK, KDS, MAP3K18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3) (Jun kinase-inhibitory kinase) (JNK/SAPK- inhibitory kinase) (Dendritic cell-derived protein kinase) (Cutaneous T-cell lymphoma tumor antigen HD-CL-09) (CTCL tumor antigen HD-CL-09) (Kinase from chicken homolog A) (hKFC-A). | |||||
|
HCDH_MOUSE
|
||||||
| NC score | 0.010668 (rank : 78) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
Domain Architecture |

|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
EMID2_MOUSE
|
||||||
| NC score | 0.007143 (rank : 79) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VF6, Q8K4P3 | Gene names | Emid2, Col26a, Col26a1, Emu2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
TRI13_MOUSE
|
||||||
| NC score | 0.005089 (rank : 80) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CYB0, Q8CEV0, Q923J0, Q925P1, Q99PQ0 | Gene names | Trim13, Rfp2 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 13 (Ret finger protein 2) (Putative tumor suppressor RFP2). | |||||