Please be patient as the page loads
|
DNJC1_MOUSE
|
||||||
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DNJC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.988253 (rank : 2) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DNJC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DNJBB_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 3) | NC score | 0.810355 (rank : 9) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
DNJBB_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 4) | NC score | 0.809659 (rank : 10) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
DNJB8_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 5) | NC score | 0.819348 (rank : 5) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 6) | NC score | 0.522782 (rank : 67) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
DNJB8_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 7) | NC score | 0.817735 (rank : 6) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DNJB6_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 8) | NC score | 0.815042 (rank : 7) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DJC16_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 9) | NC score | 0.773903 (rank : 25) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJB5_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 10) | NC score | 0.779109 (rank : 21) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
DJC16_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 11) | NC score | 0.773349 (rank : 26) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJB4_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 12) | NC score | 0.777643 (rank : 23) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DNJB5_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 13) | NC score | 0.777500 (rank : 24) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
DNJB4_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 14) | NC score | 0.778011 (rank : 22) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DNJB6_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 15) | NC score | 0.810819 (rank : 8) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DNJB7_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 16) | NC score | 0.798660 (rank : 14) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DNJB9_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 17) | NC score | 0.821796 (rank : 3) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DNJB3_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 18) | NC score | 0.808513 (rank : 11) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
DNJB9_MOUSE
|
||||||
| θ value | 5.08577e-11 (rank : 19) | NC score | 0.819525 (rank : 4) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DNJB7_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 20) | NC score | 0.802462 (rank : 12) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DJC18_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 21) | NC score | 0.785450 (rank : 19) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJBC_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 22) | NC score | 0.798752 (rank : 13) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
DNJA3_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 23) | NC score | 0.789941 (rank : 17) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJA4_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 24) | NC score | 0.764432 (rank : 34) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DNJA4_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 25) | NC score | 0.763115 (rank : 36) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
DNJB2_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 26) | NC score | 0.790185 (rank : 16) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DNJBC_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 27) | NC score | 0.790297 (rank : 15) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
DNJA1_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 28) | NC score | 0.751142 (rank : 39) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
DNJA3_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 29) | NC score | 0.787322 (rank : 18) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |
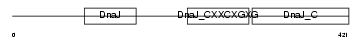
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DNJB1_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 30) | NC score | 0.766628 (rank : 32) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
DNJA1_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 31) | NC score | 0.752591 (rank : 38) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJB1_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 32) | NC score | 0.765856 (rank : 33) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DCJ14_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 33) | NC score | 0.708768 (rank : 53) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DCJ14_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 34) | NC score | 0.703257 (rank : 54) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
DJC18_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 35) | NC score | 0.763530 (rank : 35) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJC9_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 36) | NC score | 0.771957 (rank : 28) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 37) | NC score | 0.651195 (rank : 56) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 38) | NC score | 0.648714 (rank : 58) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DJC17_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 39) | NC score | 0.711742 (rank : 51) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DJC17_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 40) | NC score | 0.733351 (rank : 44) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJA2_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 41) | NC score | 0.746681 (rank : 41) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
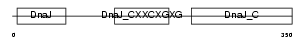
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
DNJA2_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 42) | NC score | 0.746854 (rank : 40) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
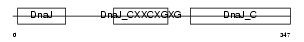
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
DNJC9_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 43) | NC score | 0.753682 (rank : 37) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
DNJBA_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 44) | NC score | 0.779414 (rank : 20) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
DNJ5B_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 45) | NC score | 0.772141 (rank : 27) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |
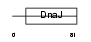
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJ5B_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 46) | NC score | 0.768015 (rank : 31) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 47) | NC score | 0.607080 (rank : 60) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 48) | NC score | 0.603827 (rank : 61) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJC5_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 49) | NC score | 0.771787 (rank : 29) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 50) | NC score | 0.771641 (rank : 30) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJBD_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 51) | NC score | 0.739449 (rank : 43) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DCJ11_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 52) | NC score | 0.725314 (rank : 47) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DCJ11_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 53) | NC score | 0.725802 (rank : 46) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DNJBD_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 54) | NC score | 0.739936 (rank : 42) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJC8_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 55) | NC score | 0.588447 (rank : 62) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 56) | NC score | 0.585396 (rank : 64) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
DNJC4_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 57) | NC score | 0.730186 (rank : 45) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
DNJC4_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 58) | NC score | 0.719529 (rank : 49) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 59) | NC score | 0.525896 (rank : 65) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 60) | NC score | 0.525656 (rank : 66) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ZCSL3_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 61) | NC score | 0.651060 (rank : 57) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
WBS18_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 62) | NC score | 0.714344 (rank : 50) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
DNJ5G_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 63) | NC score | 0.721006 (rank : 48) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
WBS18_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 64) | NC score | 0.711185 (rank : 52) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
ZCSL3_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 65) | NC score | 0.669782 (rank : 55) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 66) | NC score | 0.010654 (rank : 121) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
DJC12_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 67) | NC score | 0.619965 (rank : 59) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
CV106_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 68) | NC score | 0.093166 (rank : 76) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P0N0, Q86V14, Q96PY4, Q9NUR5, Q9Y4X9 | Gene names | C14orf106, KIAA1903 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf106 (P243). | |||||
|
ZRF1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 69) | NC score | 0.421389 (rank : 68) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
ARFP2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 70) | NC score | 0.054316 (rank : 85) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K221, Q3U3G6, Q9D7M3 | Gene names | Arfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arfaptin-2 (ADP-ribosylation factor-interacting protein 2). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 71) | NC score | 0.089368 (rank : 78) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 72) | NC score | 0.090217 (rank : 77) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
DJC12_MOUSE
|
||||||
| θ value | 0.21417 (rank : 73) | NC score | 0.587994 (rank : 63) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
MEPE_HUMAN
|
||||||
| θ value | 0.21417 (rank : 74) | NC score | 0.049704 (rank : 87) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |
|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
SF3A3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 75) | NC score | 0.040914 (rank : 89) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12874, Q15460 | Gene names | SF3A3, SAP61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3A subunit 3 (Spliceosome-associated protein 61) (SAP 61) (SF3a60). | |||||
|
ARFP2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 76) | NC score | 0.049816 (rank : 86) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53365 | Gene names | ARFIP2, POR1 | |||
|
Domain Architecture |

|
|||||
| Description | Arfaptin-2 (ADP-ribosylation factor-interacting protein 2) (Partner of RAC1) (Protein POR1). | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 77) | NC score | 0.037202 (rank : 90) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 78) | NC score | 0.075695 (rank : 82) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 79) | NC score | 0.075468 (rank : 83) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 0.47712 (rank : 80) | NC score | 0.031111 (rank : 92) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 0.62314 (rank : 81) | NC score | 0.020226 (rank : 107) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 82) | NC score | 0.020687 (rank : 105) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
MARK3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 83) | NC score | -0.000656 (rank : 136) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03141, Q3TM40, Q3V1U3, Q8C6G9, Q8R375, Q9JKE4 | Gene names | Mark3, Emk2, Mpk10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (MPK-10) (ELKL motif kinase 2). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 0.813845 (rank : 84) | NC score | 0.023583 (rank : 100) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 1.06291 (rank : 85) | NC score | 0.015918 (rank : 110) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MARK3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 86) | NC score | -0.000548 (rank : 135) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 87) | NC score | 0.011949 (rank : 118) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 1.38821 (rank : 88) | NC score | 0.009587 (rank : 123) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
CJ049_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.037034 (rank : 91) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVF2 | Gene names | C10orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf49 precursor. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.013590 (rank : 114) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.006301 (rank : 128) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
DNJCD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.392347 (rank : 70) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
PHLP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.023798 (rank : 99) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBX2, Q3TKI0 | Gene names | Pdcl | |||
|
Domain Architecture |
|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
PRC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.025670 (rank : 96) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
CDC5L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.024566 (rank : 98) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99459, Q76N46, Q99974 | Gene names | CDC5L, KIAA0432, PCDC5RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-like protein (Cdc5-like protein) (Pombe cdc5- related protein). | |||||
|
CDC5L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.024925 (rank : 97) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6A068, Q8K1J9 | Gene names | Cdc5l, Kiaa0432 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-related protein (Cdc5-like protein). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.011587 (rank : 119) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CU055_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.414889 (rank : 69) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |
|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
DHX34_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.005957 (rank : 129) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DPOLZ_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.011529 (rank : 120) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.012692 (rank : 117) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.023385 (rank : 102) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NOS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.008288 (rank : 125) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29477, O70515, O70516, Q5SXT3, Q6P6A0, Q8R410 | Gene names | Nos2, Inosl | |||
|
Domain Architecture |

|
|||||
| Description | Nitric oxide synthase, inducible (EC 1.14.13.39) (NOS type II) (Inducible NO synthase) (Inducible NOS) (iNOS) (Macrophage NOS) (MAC- NOS). | |||||
|
PHLP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.022293 (rank : 103) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13371, Q96AF1, Q9UEW7, Q9UFL0 | Gene names | PDCL | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
RM28_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.018563 (rank : 108) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13084, Q4TT39, Q96S26, Q9BQD8, Q9BR04 | Gene names | MRPL28, MAAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 39S ribosomal protein L28, mitochondrial precursor (L28mt) (MRP-L28) (Melanoma antigen p15) (Melanoma-associated antigen recognized by T lymphocytes). | |||||
|
SEBP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.023562 (rank : 101) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
UBXD8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.030712 (rank : 93) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96CS3, O94963, Q8IUF2, Q9BRP2, Q9BVM7 | Gene names | UBXD8, ETEA, KIAA0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8 (Protein ETEA). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.013493 (rank : 115) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.004786 (rank : 130) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.015565 (rank : 112) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
HXB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.001651 (rank : 132) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P0C1T1 | Gene names | Hoxb2, Hox-2.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein Hox-B2 (Hox-2.8). | |||||
|
MK07_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | -0.002531 (rank : 137) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
R51A1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.013394 (rank : 116) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.015602 (rank : 111) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
TLX3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.001315 (rank : 133) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55144 | Gene names | Tlx3, Hox11l2, Rnx, Tlx1l2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell leukemia homeobox protein 3 (Homeobox TLX-3) (Homeobox protein Hox-11L2) (Respiratory neuron homeobox protein). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.007811 (rank : 126) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.003315 (rank : 131) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.015410 (rank : 113) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.027744 (rank : 94) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.010603 (rank : 122) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.046931 (rank : 88) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.008817 (rank : 124) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
WDR60_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.016183 (rank : 109) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
DCJ15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.130036 (rank : 72) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5T4, Q5T219, Q6X963 | Gene names | DNAJC15, DNAJD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15 (Methylation-controlled J protein) (MCJ). | |||||
|
HXB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.000865 (rank : 134) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14652, P10913, P17485 | Gene names | HOXB2, HOX2H | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B2 (Hox-2H) (Hox-2.8) (K8). | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.022186 (rank : 104) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.020310 (rank : 106) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.007808 (rank : 127) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
UBXD8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.027440 (rank : 95) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3TDN2, Q3U9W2, Q80TP7, Q80W42 | Gene names | Ubxd8, Kiaa0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8. | |||||
|
AUXI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.080349 (rank : 81) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
AUXI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.070023 (rank : 84) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
CU055_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.383248 (rank : 71) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NX36 | Gene names | C21orf55 | |||
|
Domain Architecture |
|
|||||
| Description | J domain-containing protein C21orf55. | |||||
|
DCJ15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.095879 (rank : 75) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q78YY6 | Gene names | Dnajc15, Dnajd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15. | |||||
|
SACS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.085732 (rank : 80) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.086338 (rank : 79) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
TIM14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.122642 (rank : 73) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96DA6 | Gene names | DNAJC19, TIM14, TIMM14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
TIM14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.122217 (rank : 74) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CQV7, Q8R1N1, Q9D896 | Gene names | Dnajc19, Tim14, Timm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
DNJC1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DNJC1_HUMAN
|
||||||
| NC score | 0.988253 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DNJB9_HUMAN
|
||||||
| NC score | 0.821796 (rank : 3) | θ value | 1.74796e-11 (rank : 17) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DNJB9_MOUSE
|
||||||
| NC score | 0.819525 (rank : 4) | θ value | 5.08577e-11 (rank : 19) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DNJB8_MOUSE
|
||||||
| NC score | 0.819348 (rank : 5) | θ value | 1.86753e-13 (rank : 5) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJB8_HUMAN
|
||||||
| NC score | 0.817735 (rank : 6) | θ value | 7.09661e-13 (rank : 7) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DNJB6_HUMAN
|
||||||
| NC score | 0.815042 (rank : 7) | θ value | 2.0648e-12 (rank : 8) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJB6_MOUSE
|
||||||
| NC score | 0.810819 (rank : 8) | θ value | 1.02475e-11 (rank : 15) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DNJBB_HUMAN
|
||||||
| NC score | 0.810355 (rank : 9) | θ value | 3.76295e-14 (rank : 3) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
DNJBB_MOUSE
|
||||||
| NC score | 0.809659 (rank : 10) | θ value | 6.41864e-14 (rank : 4) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
DNJB3_MOUSE
|
||||||
| NC score | 0.808513 (rank : 11) | θ value | 2.28291e-11 (rank : 18) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
DNJB7_MOUSE
|
||||||
| NC score | 0.802462 (rank : 12) | θ value | 6.64225e-11 (rank : 20) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DNJBC_MOUSE
|
||||||
| NC score | 0.798752 (rank : 13) | θ value | 1.9326e-10 (rank : 22) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
DNJB7_HUMAN
|
||||||
| NC score | 0.798660 (rank : 14) | θ value | 1.33837e-11 (rank : 16) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DNJBC_HUMAN
|
||||||
| NC score | 0.790297 (rank : 15) | θ value | 5.62301e-10 (rank : 27) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
DNJB2_HUMAN
|
||||||
| NC score | 0.790185 (rank : 16) | θ value | 5.62301e-10 (rank : 26) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DNJA3_MOUSE
|
||||||
| NC score | 0.789941 (rank : 17) | θ value | 3.29651e-10 (rank : 23) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJA3_HUMAN
|
||||||
| NC score | 0.787322 (rank : 18) | θ value | 9.59137e-10 (rank : 29) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DJC18_HUMAN
|
||||||
| NC score | 0.785450 (rank : 19) | θ value | 1.47974e-10 (rank : 21) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJBA_MOUSE
|
||||||
| NC score | 0.779414 (rank : 20) | θ value | 5.26297e-08 (rank : 44) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
DNJB5_MOUSE
|
||||||
| NC score | 0.779109 (rank : 21) | θ value | 4.59992e-12 (rank : 10) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
DNJB4_MOUSE
|
||||||
| NC score | 0.778011 (rank : 22) | θ value | 1.02475e-11 (rank : 14) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DNJB4_HUMAN
|
||||||
| NC score | 0.777643 (rank : 23) | θ value | 6.00763e-12 (rank : 12) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DNJB5_HUMAN
|
||||||
| NC score | 0.777500 (rank : 24) | θ value | 7.84624e-12 (rank : 13) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
DJC16_HUMAN
|
||||||
| NC score | 0.773903 (rank : 25) | θ value | 2.69671e-12 (rank : 9) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DJC16_MOUSE
|
||||||
| NC score | 0.773349 (rank : 26) | θ value | 6.00763e-12 (rank : 11) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJ5B_HUMAN
|
||||||
| NC score | 0.772141 (rank : 27) | θ value | 8.97725e-08 (rank : 45) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJC9_HUMAN
|
||||||
| NC score | 0.771957 (rank : 28) | θ value | 8.11959e-09 (rank : 36) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
DNJC5_HUMAN
|
||||||
| NC score | 0.771787 (rank : 29) | θ value | 2.61198e-07 (rank : 49) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| NC score | 0.771641 (rank : 30) | θ value | 2.61198e-07 (rank : 50) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJ5B_MOUSE
|
||||||
| NC score | 0.768015 (rank : 31) | θ value | 1.99992e-07 (rank : 46) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJB1_HUMAN
|
||||||
| NC score | 0.766628 (rank : 32) | θ value | 9.59137e-10 (rank : 30) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
DNJB1_MOUSE
|
||||||
| NC score | 0.765856 (rank : 33) | θ value | 1.25267e-09 (rank : 32) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DNJA4_HUMAN
|
||||||
| NC score | 0.764432 (rank : 34) | θ value | 5.62301e-10 (rank : 24) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DJC18_MOUSE
|
||||||
| NC score | 0.763530 (rank : 35) | θ value | 6.21693e-09 (rank : 35) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJA4_MOUSE
|
||||||
| NC score | 0.763115 (rank : 36) | θ value | 5.62301e-10 (rank : 25) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
DNJC9_MOUSE
|
||||||
| NC score | 0.753682 (rank : 37) | θ value | 4.0297e-08 (rank : 43) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
DNJA1_HUMAN
|
||||||
| NC score | 0.752591 (rank : 38) | θ value | 1.25267e-09 (rank : 31) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJA1_MOUSE
|
||||||
| NC score | 0.751142 (rank : 39) | θ value | 9.59137e-10 (rank : 28) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
DNJA2_MOUSE
|
||||||
| NC score | 0.746854 (rank : 40) | θ value | 4.0297e-08 (rank : 42) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
DNJA2_HUMAN
|
||||||
| NC score | 0.746681 (rank : 41) | θ value | 4.0297e-08 (rank : 41) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
DNJBD_HUMAN
|
||||||
| NC score | 0.739936 (rank : 42) | θ value | 7.59969e-07 (rank : 54) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJBD_MOUSE
|
||||||
| NC score | 0.739449 (rank : 43) | θ value | 4.45536e-07 (rank : 51) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DJC17_MOUSE
|
||||||
| NC score | 0.733351 (rank : 44) | θ value | 4.0297e-08 (rank : 40) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJC4_HUMAN
|
||||||
| NC score | 0.730186 (rank : 45) | θ value | 3.77169e-06 (rank : 57) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
DCJ11_MOUSE
|
||||||
| NC score | 0.725802 (rank : 46) | θ value | 5.81887e-07 (rank : 53) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DCJ11_HUMAN
|
||||||
| NC score | 0.725314 (rank : 47) | θ value | 5.81887e-07 (rank : 52) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DNJ5G_HUMAN
|
||||||
| NC score | 0.721006 (rank : 48) | θ value | 0.00298849 (rank : 63) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
DNJC4_MOUSE
|
||||||
| NC score | 0.719529 (rank : 49) | θ value | 4.92598e-06 (rank : 58) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
WBS18_MOUSE
|
||||||
| NC score | 0.714344 (rank : 50) | θ value | 0.00175202 (rank : 62) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
DJC17_HUMAN
|
||||||
| NC score | 0.711742 (rank : 51) | θ value | 4.0297e-08 (rank : 39) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
WBS18_HUMAN
|
||||||
| NC score | 0.711185 (rank : 52) | θ value | 0.00509761 (rank : 64) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
DCJ14_MOUSE
|
||||||
| NC score | 0.708768 (rank : 53) | θ value | 1.63604e-09 (rank : 33) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DCJ14_HUMAN
|
||||||
| NC score | 0.703257 (rank : 54) | θ value | 3.64472e-09 (rank : 34) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
ZCSL3_HUMAN
|
||||||
| NC score | 0.669782 (rank : 55) | θ value | 0.00665767 (rank : 65) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.651195 (rank : 56) | θ value | 3.08544e-08 (rank : 37) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
ZCSL3_MOUSE
|
||||||
| NC score | 0.651060 (rank : 57) | θ value | 0.000461057 (rank : 61) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.648714 (rank : 58) | θ value | 3.08544e-08 (rank : 38) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DJC12_HUMAN
|
||||||
| NC score | 0.619965 (rank : 59) | θ value | 0.0431538 (rank : 67) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.607080 (rank : 60) | θ value | 1.99992e-07 (rank : 47) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.603827 (rank : 61) | θ value | 1.99992e-07 (rank : 48) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJC8_HUMAN
|
||||||
| NC score | 0.588447 (rank : 62) | θ value | 2.88788e-06 (rank : 55) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DJC12_MOUSE
|
||||||
| NC score | 0.587994 (rank : 63) | θ value | 0.21417 (rank : 73) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DNJC8_MOUSE
|
||||||
| NC score | 0.585396 (rank : 64) | θ value | 2.88788e-06 (rank : 56) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.525896 (rank : 65) | θ value | 4.92598e-06 (rank : 59) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.525656 (rank : 66) | θ value | 4.92598e-06 (rank : 60) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.522782 (rank : 67) | θ value | 5.43371e-13 (rank : 6) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
ZRF1_MOUSE
|
||||||
| NC score | 0.421389 (rank : 68) | θ value | 0.0563607 (rank : 69) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
CU055_MOUSE
|
||||||
| NC score | 0.414889 (rank : 69) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |

|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
DNJCD_HUMAN
|
||||||
| NC score | 0.392347 (rank : 70) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
CU055_HUMAN
|
||||||
| NC score | 0.383248 (rank : 71) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NX36 | Gene names | C21orf55 | |||
|
Domain Architecture |

|
|||||
| Description | J domain-containing protein C21orf55. | |||||
|
DCJ15_HUMAN
|
||||||
| NC score | 0.130036 (rank : 72) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5T4, Q5T219, Q6X963 | Gene names | DNAJC15, DNAJD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15 (Methylation-controlled J protein) (MCJ). | |||||
|
TIM14_HUMAN
|
||||||
| NC score | 0.122642 (rank : 73) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96DA6 | Gene names | DNAJC19, TIM14, TIMM14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
TIM14_MOUSE
|
||||||
| NC score | 0.122217 (rank : 74) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CQV7, Q8R1N1, Q9D896 | Gene names | Dnajc19, Tim14, Timm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
DCJ15_MOUSE
|
||||||
| NC score | 0.095879 (rank : 75) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q78YY6 | Gene names | Dnajc15, Dnajd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15. | |||||
|
CV106_HUMAN
|
||||||
| NC score | 0.093166 (rank : 76) | θ value | 0.0563607 (rank : 68) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P0N0, Q86V14, Q96PY4, Q9NUR5, Q9Y4X9 | Gene names | C14orf106, KIAA1903 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf106 (P243). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.090217 (rank : 77) | θ value | 0.125558 (rank : 72) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.089368 (rank : 78) | θ value | 0.0961366 (rank : 71) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.086338 (rank : 79) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.085732 (rank : 80) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
AUXI_HUMAN
|
||||||
| NC score | 0.080349 (rank : 81) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.075695 (rank : 82) | θ value | 0.365318 (rank : 78) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.075468 (rank : 83) | θ value | 0.365318 (rank : 79) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
AUXI_MOUSE
|
||||||
| NC score | 0.070023 (rank : 84) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
ARFP2_MOUSE
|
||||||
| NC score | 0.054316 (rank : 85) | θ value | 0.0961366 (rank : 70) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K221, Q3U3G6, Q9D7M3 | Gene names | Arfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arfaptin-2 (ADP-ribosylation factor-interacting protein 2). | |||||
|
ARFP2_HUMAN
|
||||||
| NC score | 0.049816 (rank : 86) | θ value | 0.365318 (rank : 76) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53365 | Gene names | ARFIP2, POR1 | |||
|
Domain Architecture |

|
|||||
| Description | Arfaptin-2 (ADP-ribosylation factor-interacting protein 2) (Partner of RAC1) (Protein POR1). | |||||
|
MEPE_HUMAN
|
||||||
| NC score | 0.049704 (rank : 87) | θ value | 0.21417 (rank : 74) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |

|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.046931 (rank : 88) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
SF3A3_HUMAN
|
||||||
| NC score | 0.040914 (rank : 89) | θ value | 0.279714 (rank : 75) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12874, Q15460 | Gene names | SF3A3, SAP61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3A subunit 3 (Spliceosome-associated protein 61) (SAP 61) (SF3a60). | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.037202 (rank : 90) | θ value | 0.365318 (rank : 77) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CJ049_HUMAN
|
||||||
| NC score | 0.037034 (rank : 91) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVF2 | Gene names | C10orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf49 precursor. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.031111 (rank : 92) | θ value | 0.47712 (rank : 80) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
UBXD8_HUMAN
|
||||||
| NC score | 0.030712 (rank : 93) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96CS3, O94963, Q8IUF2, Q9BRP2, Q9BVM7 | Gene names | UBXD8, ETEA, KIAA0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8 (Protein ETEA). | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.027744 (rank : 94) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
UBXD8_MOUSE
|
||||||
| NC score | 0.027440 (rank : 95) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3TDN2, Q3U9W2, Q80TP7, Q80W42 | Gene names | Ubxd8, Kiaa0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8. | |||||
|
PRC1_MOUSE
|
||||||
| NC score | 0.025670 (rank : 96) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
CDC5L_MOUSE
|
||||||
| NC score | 0.024925 (rank : 97) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6A068, Q8K1J9 | Gene names | Cdc5l, Kiaa0432 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-related protein (Cdc5-like protein). | |||||
|
CDC5L_HUMAN
|
||||||
| NC score | 0.024566 (rank : 98) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99459, Q76N46, Q99974 | Gene names | CDC5L, KIAA0432, PCDC5RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-like protein (Cdc5-like protein) (Pombe cdc5- related protein). | |||||
|
PHLP_MOUSE
|
||||||
| NC score | 0.023798 (rank : 99) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBX2, Q3TKI0 | Gene names | Pdcl | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.023583 (rank : 100) | θ value | 0.813845 (rank : 84) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
SEBP2_HUMAN
|
||||||
| NC score | 0.023562 (rank : 101) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.023385 (rank : 102) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PHLP_HUMAN
|
||||||
| NC score | 0.022293 (rank : 103) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13371, Q96AF1, Q9UEW7, Q9UFL0 | Gene names | PDCL | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.022186 (rank : 104) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.020687 (rank : 105) | θ value | 0.813845 (rank : 82) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.020310 (rank : 106) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.020226 (rank : 107) | θ value | 0.62314 (rank : 81) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
RM28_HUMAN
|
||||||
| NC score | 0.018563 (rank : 108) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13084, Q4TT39, Q96S26, Q9BQD8, Q9BR04 | Gene names | MRPL28, MAAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 39S ribosomal protein L28, mitochondrial precursor (L28mt) (MRP-L28) (Melanoma antigen p15) (Melanoma-associated antigen recognized by T lymphocytes). | |||||
|
WDR60_HUMAN
|
||||||
| NC score | 0.016183 (rank : 109) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.015918 (rank : 110) | θ value | 1.06291 (rank : 85) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.015602 (rank : 111) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.015565 (rank : 112) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.015410 (rank : 113) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.013590 (rank : 114) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.013493 (rank : 115) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.013394 (rank : 116) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.012692 (rank : 117) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.011949 (rank : 118) | θ value | 1.38821 (rank : 87) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.011587 (rank : 119) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DPOLZ_HUMAN
|
||||||
| NC score | 0.011529 (rank : 120) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.010654 (rank : 121) | θ value | 0.0193708 (rank : 66) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.010603 (rank : 122) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.009587 (rank : 123) | θ value | 1.38821 (rank : 88) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.008817 (rank : 124) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
NOS2_MOUSE
|
||||||
| NC score | 0.008288 (rank : 125) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29477, O70515, O70516, Q5SXT3, Q6P6A0, Q8R410 | Gene names | Nos2, Inosl | |||
|
Domain Architecture |

|
|||||
| Description | Nitric oxide synthase, inducible (EC 1.14.13.39) (NOS type II) (Inducible NO synthase) (Inducible NOS) (iNOS) (Macrophage NOS) (MAC- NOS). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.007811 (rank : 126) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.007808 (rank : 127) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.006301 (rank : 128) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
DHX34_MOUSE
|
||||||
| NC score | 0.005957 (rank : 129) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.004786 (rank : 130) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.003315 (rank : 131) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
HXB2_MOUSE
|
||||||
| NC score | 0.001651 (rank : 132) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P0C1T1 | Gene names | Hoxb2, Hox-2.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein Hox-B2 (Hox-2.8). | |||||
|
TLX3_MOUSE
|
||||||
| NC score | 0.001315 (rank : 133) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55144 | Gene names | Tlx3, Hox11l2, Rnx, Tlx1l2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell leukemia homeobox protein 3 (Homeobox TLX-3) (Homeobox protein Hox-11L2) (Respiratory neuron homeobox protein). | |||||
|
HXB2_HUMAN
|
||||||
| NC score | 0.000865 (rank : 134) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14652, P10913, P17485 | Gene names | HOXB2, HOX2H | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B2 (Hox-2H) (Hox-2.8) (K8). | |||||
|
MARK3_HUMAN
|
||||||
| NC score | -0.000548 (rank : 135) | θ value | 1.38821 (rank : 86) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
MARK3_MOUSE
|
||||||
| NC score | -0.000656 (rank : 136) | θ value | 0.813845 (rank : 83) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03141, Q3TM40, Q3V1U3, Q8C6G9, Q8R375, Q9JKE4 | Gene names | Mark3, Emk2, Mpk10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (MPK-10) (ELKL motif kinase 2). | |||||
|
MK07_HUMAN
|
||||||
| NC score | -0.002531 (rank : 137) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||