Please be patient as the page loads
|
AP3D1_MOUSE
|
||||||
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AP3D1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.959854 (rank : 2) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | 1.12253e-42 (rank : 3) | NC score | 0.796802 (rank : 5) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | 1.91475e-42 (rank : 4) | NC score | 0.794022 (rank : 6) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP4E1_MOUSE
|
||||||
| θ value | 8.04462e-41 (rank : 5) | NC score | 0.809541 (rank : 4) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_HUMAN
|
||||||
| θ value | 3.99248e-40 (rank : 6) | NC score | 0.809650 (rank : 3) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP2A2_MOUSE
|
||||||
| θ value | 1.57e-36 (rank : 7) | NC score | 0.774125 (rank : 10) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| θ value | 2.05049e-36 (rank : 8) | NC score | 0.774828 (rank : 9) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | 2.26708e-35 (rank : 9) | NC score | 0.783636 (rank : 7) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_HUMAN
|
||||||
| θ value | 1.46948e-34 (rank : 10) | NC score | 0.781064 (rank : 8) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP2A1_MOUSE
|
||||||
| θ value | 1.79631e-32 (rank : 11) | NC score | 0.761273 (rank : 12) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_HUMAN
|
||||||
| θ value | 2.34606e-32 (rank : 12) | NC score | 0.765714 (rank : 11) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP1B1_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 13) | NC score | 0.583555 (rank : 14) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP1B1_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 14) | NC score | 0.584853 (rank : 13) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP2B1_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 15) | NC score | 0.575650 (rank : 16) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 16) | NC score | 0.575667 (rank : 15) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
COPB_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 17) | NC score | 0.518816 (rank : 17) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPB_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 18) | NC score | 0.518435 (rank : 18) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
AP4B1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 19) | NC score | 0.381478 (rank : 21) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6B7 | Gene names | AP4B1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta 1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 20) | NC score | 0.379357 (rank : 22) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 21) | NC score | 0.384127 (rank : 20) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP4B1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 22) | NC score | 0.396952 (rank : 19) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WV76 | Gene names | Ap4b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta-1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 23) | NC score | 0.345546 (rank : 23) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 24) | NC score | 0.344717 (rank : 24) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 25) | NC score | 0.093781 (rank : 33) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 26) | NC score | 0.090418 (rank : 36) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
AFF2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 27) | NC score | 0.092064 (rank : 34) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 28) | NC score | 0.081125 (rank : 42) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.082052 (rank : 40) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 30) | NC score | 0.061867 (rank : 54) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 31) | NC score | 0.017919 (rank : 134) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 32) | NC score | 0.067735 (rank : 48) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
TTF1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 33) | NC score | 0.103927 (rank : 28) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15361, Q4VXF3, Q58EY2, Q6P5T5 | Gene names | TTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (RNA polymerase I termination factor). | |||||
|
COPG_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 34) | NC score | 0.204785 (rank : 27) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 35) | NC score | 0.036967 (rank : 83) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
COPG2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 36) | NC score | 0.233638 (rank : 25) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXK3 | Gene names | Copg2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 37) | NC score | 0.087620 (rank : 38) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 38) | NC score | 0.019669 (rank : 123) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 39) | NC score | 0.089306 (rank : 37) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.033094 (rank : 90) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | 0.125558 (rank : 41) | NC score | 0.031806 (rank : 94) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
SPAG6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 42) | NC score | 0.072644 (rank : 46) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
COPG2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 43) | NC score | 0.222538 (rank : 26) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBF2 | Gene names | COPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
XRCC4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 44) | NC score | 0.050837 (rank : 71) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924T3, Q8BKC9, Q8BU02, Q9D6U6 | Gene names | Xrcc4 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
IF4E3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 45) | NC score | 0.051588 (rank : 69) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60573, O75349 | Gene names | EIF4EL3 | |||
|
Domain Architecture |
|
|||||
| Description | Eukaryotic translation initiation factor 4E type 3 (eIF4E type 3) (eIF-4E type 3) (mRNA cap-binding protein type 3) (Eukaryotic translation initiation factor 4E-like 3) (Eukaryotic translation initiation factor 4E homologous protein) (mRNA cap-binding protein 4EHP) (eIF4E-like protein 4E-LP). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 46) | NC score | 0.048841 (rank : 72) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 0.21417 (rank : 47) | NC score | 0.077019 (rank : 43) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.090456 (rank : 35) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.076522 (rank : 44) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
A4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.034700 (rank : 86) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.062319 (rank : 53) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.033579 (rank : 87) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
STOX1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.046886 (rank : 73) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
MA2A2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.022887 (rank : 108) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49641, Q13754 | Gene names | MAN2A2, MANA2X | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase IIx (EC 3.2.1.114) (Mannosyl-oligosaccharide 1,3- 1,6-alpha-mannosidase) (MAN IIx) (Mannosidase alpha class 2A member 2). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.024328 (rank : 101) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.024262 (rank : 102) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.075722 (rank : 45) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.067159 (rank : 49) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
A4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.033249 (rank : 88) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
RIT1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.006142 (rank : 157) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92963, O00646, O00720 | Gene names | RIT1, RIBB, RIT, ROC1 | |||
|
Domain Architecture |
|
|||||
| Description | GTP-binding protein Rit1 (Ras-like protein expressed in many tissues) (Ras-like without CAAX protein 1). | |||||
|
CF010_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.059551 (rank : 56) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
IF4E3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.045571 (rank : 74) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMB3, O88503, Q3UCK2 | Gene names | Eif4el3 | |||
|
Domain Architecture |
|
|||||
| Description | Eukaryotic translation initiation factor 4E type 3 (eIF4E type 3) (eIF-4E type 3) (mRNA cap-binding protein type 3) (Eukaryotic translation initiation factor 4E-like 3) (eIF4E-like protein 4E-LP). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.040365 (rank : 76) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
TR150_HUMAN
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.060871 (rank : 55) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.020738 (rank : 120) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.023658 (rank : 106) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.023478 (rank : 107) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.018481 (rank : 132) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.036459 (rank : 84) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.030240 (rank : 95) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.009852 (rank : 155) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
BCAS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.021712 (rank : 117) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CCN5, Q8CC16, Q9EPX3 | Gene names | Bcas3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 3 homolog (K20D4). | |||||
|
DJC12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.021720 (rank : 116) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DNJCD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.034815 (rank : 85) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.013804 (rank : 147) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.012237 (rank : 151) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TBC14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.023747 (rank : 105) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGA2, Q8CHA5 | Gene names | Tbc1d14, D5Ertd110e, Kiaa1322 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 14. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.039136 (rank : 78) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ZN346_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.023970 (rank : 104) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UL40, Q68CV9, Q6ZMW1 | Gene names | ZNF346, JAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 346 (Just another zinc finger protein). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.021428 (rank : 118) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.028077 (rank : 96) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.026850 (rank : 98) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.014944 (rank : 144) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
I20RB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.022697 (rank : 109) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UXL0, Q6P438, Q8TAJ7 | Gene names | IL20RB, DIRS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-20 receptor beta chain precursor (IL-20R-beta) (IL-20R2). | |||||
|
NEBU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.012306 (rank : 150) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |
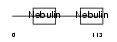
|
|||||
| Description | Nebulin. | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.037243 (rank : 81) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.032178 (rank : 93) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.032695 (rank : 91) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.033204 (rank : 89) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ZN688_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | -0.002881 (rank : 161) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
ANGP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.008306 (rank : 156) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15123, Q9NRR7, Q9P2Y7 | Gene names | ANGPT2 | |||
|
Domain Architecture |
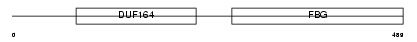
|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.038431 (rank : 79) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
CCD55_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.039331 (rank : 77) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.036999 (rank : 82) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.019403 (rank : 127) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.044412 (rank : 75) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.064968 (rank : 50) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RIT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.003859 (rank : 159) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70426 | Gene names | Rit1, Rit | |||
|
Domain Architecture |
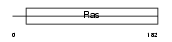
|
|||||
| Description | GTP-binding protein Rit1 (Ras-like protein expressed in many tissues) (Ras-like without CAAX protein 1). | |||||
|
SYMPK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.024016 (rank : 103) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.019354 (rank : 128) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.032575 (rank : 92) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
DYN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.009937 (rank : 154) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
ESPL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.017857 (rank : 135) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14674 | Gene names | ESPL1, ESP1, KIAA0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.018804 (rank : 130) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
JOSD3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.021870 (rank : 115) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D4V4, Q149X7, Q8C7I5, Q9CZG0, Q9D5U6 | Gene names | Josd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein JOSD3. | |||||
|
NBN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.022179 (rank : 111) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R207, O88981, Q3UY57, Q811I6, Q8CCY0, Q9R1X1 | Gene names | Nbn, Nbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1 homolog) (Cell cycle regulatory protein p95). | |||||
|
PCTL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.020815 (rank : 119) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |
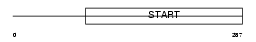
|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
PHC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.019469 (rank : 125) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.016327 (rank : 138) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.025811 (rank : 99) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.037755 (rank : 80) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.052161 (rank : 66) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
XRN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.017954 (rank : 133) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97789, O35651, P97790, Q3TE44, Q3TRE9, Q3UQL5 | Gene names | Xrn1, Dhm2, Exo, Sep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (mXRN1) (Strand-exchange protein 1 homolog) (Protein Dhm2). | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.052676 (rank : 65) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.015571 (rank : 140) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.027328 (rank : 97) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.016482 (rank : 137) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
CEBPB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.012824 (rank : 149) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
CF081_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.019489 (rank : 124) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80X86, Q9D564, Q9DAN7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81 homolog. | |||||
|
EVPL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.021934 (rank : 114) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
IF39_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.022164 (rank : 112) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.022554 (rank : 110) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
KLC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.020210 (rank : 121) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.018635 (rank : 131) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.019140 (rank : 129) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NPHP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.013006 (rank : 148) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.013814 (rank : 146) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
O5AT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | -0.003266 (rank : 163) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHC5 | Gene names | OR5AT1 | |||
|
Domain Architecture |
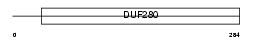
|
|||||
| Description | Olfactory receptor 5AT1. | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.017090 (rank : 136) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
TR240_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.015145 (rank : 141) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHV7, O60334 | Gene names | THRAP1, ARC250, KIAA0593, TRAP240 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor-associated protein complex 240 kDa component (Trap240) (Thyroid hormone receptor-associated protein 1) (Vitamin D3 receptor-interacting protein complex component DRIP250) (DRIP 250) (Activator-recruited cofactor 250 kDa component) (ARC250). | |||||
|
ZN406_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | -0.003089 (rank : 162) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TS63 | Gene names | Znf406, Zfp406 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406. | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.003390 (rank : 160) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.015620 (rank : 139) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.015010 (rank : 143) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
K1024_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.014552 (rank : 145) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPX6 | Gene names | KIAA1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024. | |||||
|
K1024_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.015111 (rank : 142) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3V7, Q69ZS9 | Gene names | Kiaa1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024 (Protein DD1). | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.004976 (rank : 158) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
NONO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.010947 (rank : 152) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |
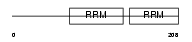
|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
NONO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.010795 (rank : 153) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.064170 (rank : 51) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.022059 (rank : 113) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.020177 (rank : 122) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZN292_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.019457 (rank : 126) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.025761 (rank : 100) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.058560 (rank : 59) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.063566 (rank : 52) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
GGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.085778 (rank : 39) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.081639 (rank : 41) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.096689 (rank : 30) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.100562 (rank : 29) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.093783 (rank : 32) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.093953 (rank : 31) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.054267 (rank : 62) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.054217 (rank : 63) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.059214 (rank : 58) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.051729 (rank : 68) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
RRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.053517 (rank : 64) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.059400 (rank : 57) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.055337 (rank : 60) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.052005 (rank : 67) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.070631 (rank : 47) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.051508 (rank : 70) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.055273 (rank : 61) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.959854 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP4E1_HUMAN
|
||||||
| NC score | 0.809650 (rank : 3) | θ value | 3.99248e-40 (rank : 6) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_MOUSE
|
||||||
| NC score | 0.809541 (rank : 4) | θ value | 8.04462e-41 (rank : 5) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.796802 (rank : 5) | θ value | 1.12253e-42 (rank : 3) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.794022 (rank : 6) | θ value | 1.91475e-42 (rank : 4) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.783636 (rank : 7) | θ value | 2.26708e-35 (rank : 9) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_HUMAN
|
||||||
| NC score | 0.781064 (rank : 8) | θ value | 1.46948e-34 (rank : 10) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP2A2_HUMAN
|
||||||
| NC score | 0.774828 (rank : 9) | θ value | 2.05049e-36 (rank : 8) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| NC score | 0.774125 (rank : 10) | θ value | 1.57e-36 (rank : 7) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP2A1_HUMAN
|
||||||
| NC score | 0.765714 (rank : 11) | θ value | 2.34606e-32 (rank : 12) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| NC score | 0.761273 (rank : 12) | θ value | 1.79631e-32 (rank : 11) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP1B1_MOUSE
|
||||||
| NC score | 0.584853 (rank : 13) | θ value | 2.5182e-18 (rank : 14) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP1B1_HUMAN
|
||||||
| NC score | 0.583555 (rank : 14) | θ value | 2.5182e-18 (rank : 13) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP2B1_MOUSE
|
||||||
| NC score | 0.575667 (rank : 15) | θ value | 2.13179e-17 (rank : 16) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_HUMAN
|
||||||
| NC score | 0.575650 (rank : 16) | θ value | 2.13179e-17 (rank : 15) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
COPB_HUMAN
|
||||||
| NC score | 0.518816 (rank : 17) | θ value | 5.25075e-16 (rank : 17) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPB_MOUSE
|
||||||
| NC score | 0.518435 (rank : 18) | θ value | 1.16975e-15 (rank : 18) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
AP4B1_MOUSE
|
||||||
| NC score | 0.396952 (rank : 19) | θ value | 0.00020696 (rank : 22) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WV76 | Gene names | Ap4b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta-1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.384127 (rank : 20) | θ value | 0.00020696 (rank : 21) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP4B1_HUMAN
|
||||||
| NC score | 0.381478 (rank : 21) | θ value | 7.1131e-05 (rank : 19) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6B7 | Gene names | AP4B1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta 1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.379357 (rank : 22) | θ value | 0.000158464 (rank : 20) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.345546 (rank : 23) | θ value | 0.00134147 (rank : 23) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.344717 (rank : 24) | θ value | 0.00134147 (rank : 24) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
COPG2_MOUSE
|
||||||
| NC score | 0.233638 (rank : 25) | θ value | 0.0563607 (rank : 36) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXK3 | Gene names | Copg2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG2_HUMAN
|
||||||
| NC score | 0.222538 (rank : 26) | θ value | 0.163984 (rank : 43) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBF2 | Gene names | COPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG_HUMAN
|
||||||
| NC score | 0.204785 (rank : 27) | θ value | 0.0431538 (rank : 34) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
TTF1_HUMAN
|
||||||
| NC score | 0.103927 (rank : 28) | θ value | 0.0330416 (rank : 33) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15361, Q4VXF3, Q58EY2, Q6P5T5 | Gene names | TTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (RNA polymerase I termination factor). | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.100562 (rank : 29) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.096689 (rank : 30) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.093953 (rank : 31) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.093783 (rank : 32) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.093781 (rank : 33) | θ value | 0.00228821 (rank : 25) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
AFF2_MOUSE
|
||||||
| NC score | 0.092064 (rank : 34) | θ value | 0.0148317 (rank : 27) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.090456 (rank : 35) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.090418 (rank : 36) | θ value | 0.00665767 (rank : 26) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.089306 (rank : 37) | θ value | 0.0961366 (rank : 39) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.087620 (rank : 38) | θ value | 0.0563607 (rank : 37) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.085778 (rank : 39) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.082052 (rank : 40) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
GGA1_MOUSE
|
||||||
| NC score | 0.081639 (rank : 41) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.081125 (rank : 42) | θ value | 0.0148317 (rank : 28) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.077019 (rank : 43) | θ value | 0.21417 (rank : 47) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.076522 (rank : 44) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.075722 (rank : 45) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
SPAG6_HUMAN
|
||||||
| NC score | 0.072644 (rank : 46) | θ value | 0.125558 (rank : 42) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.070631 (rank : 47) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.067735 (rank : 48) | θ value | 0.0330416 (rank : 32) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.067159 (rank : 49) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.064968 (rank : 50) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.064170 (rank : 51) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.063566 (rank : 52) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.062319 (rank : 53) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.061867 (rank : 54) | θ value | 0.0252991 (rank : 30) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TR150_HUMAN
|
||||||
| NC score | 0.060871 (rank : 55) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.059551 (rank : 56) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.059400 (rank : 57) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.059214 (rank : 58) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.058560 (rank : 59) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.055337 (rank : 60) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.055273 (rank : 61) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.054267 (rank : 62) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.054217 (rank : 63) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RRP5_HUMAN
|
||||||
| NC score | 0.053517 (rank : 64) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.052676 (rank : 65) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.052161 (rank : 66) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.052005 (rank : 67) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.051729 (rank : 68) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
IF4E3_HUMAN
|
||||||
| NC score | 0.051588 (rank : 69) | θ value | 0.21417 (rank : 45) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60573, O75349 | Gene names | EIF4EL3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4E type 3 (eIF4E type 3) (eIF-4E type 3) (mRNA cap-binding protein type 3) (Eukaryotic translation initiation factor 4E-like 3) (Eukaryotic translation initiation factor 4E homologous protein) (mRNA cap-binding protein 4EHP) (eIF4E-like protein 4E-LP). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.051508 (rank : 70) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
XRCC4_MOUSE
|
||||||
| NC score | 0.050837 (rank : 71) | θ value | 0.163984 (rank : 44) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924T3, Q8BKC9, Q8BU02, Q9D6U6 | Gene names | Xrcc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.048841 (rank : 72) | θ value | 0.21417 (rank : 46) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
STOX1_HUMAN
|
||||||
| NC score | 0.046886 (rank : 73) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
IF4E3_MOUSE
|
||||||
| NC score | 0.045571 (rank : 74) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMB3, O88503, Q3UCK2 | Gene names | Eif4el3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4E type 3 (eIF4E type 3) (eIF-4E type 3) (mRNA cap-binding protein type 3) (Eukaryotic translation initiation factor 4E-like 3) (eIF4E-like protein 4E-LP). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.044412 (rank : 75) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.040365 (rank : 76) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.039331 (rank : 77) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.039136 (rank : 78) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.038431 (rank : 79) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.037755 (rank : 80) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.037243 (rank : 81) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.036999 (rank : 82) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.036967 (rank : 83) | θ value | 0.0563607 (rank : 35) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.036459 (rank : 84) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
DNJCD_HUMAN
|
||||||
| NC score | 0.034815 (rank : 85) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.034700 (rank : 86) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.033579 (rank : 87) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.033249 (rank : 88) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.033204 (rank : 89) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.033094 (rank : 90) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.032695 (rank : 91) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.032575 (rank : 92) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.032178 (rank : 93) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.031806 (rank : 94) | θ value | 0.125558 (rank : 41) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.030240 (rank : 95) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.028077 (rank : 96) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.027328 (rank : 97) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.026850 (rank : 98) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.025811 (rank : 99) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.025761 (rank : 100) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.024328 (rank : 101) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.024262 (rank : 102) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SYMPK_MOUSE
|
||||||
| NC score | 0.024016 (rank : 103) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
ZN346_HUMAN
|
||||||
| NC score | 0.023970 (rank : 104) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UL40, Q68CV9, Q6ZMW1 | Gene names | ZNF346, JAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 346 (Just another zinc finger protein). | |||||
|
TBC14_MOUSE
|
||||||
| NC score | 0.023747 (rank : 105) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGA2, Q8CHA5 | Gene names | Tbc1d14, D5Ertd110e, Kiaa1322 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 14. | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.023658 (rank : 106) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.023478 (rank : 107) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MA2A2_HUMAN
|
||||||
| NC score | 0.022887 (rank : 108) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49641, Q13754 | Gene names | MAN2A2, MANA2X | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase IIx (EC 3.2.1.114) (Mannosyl-oligosaccharide 1,3- 1,6-alpha-mannosidase) (MAN IIx) (Mannosidase alpha class 2A member 2). | |||||
|
I20RB_HUMAN
|
||||||
| NC score | 0.022697 (rank : 109) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UXL0, Q6P438, Q8TAJ7 | Gene names | IL20RB, DIRS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-20 receptor beta chain precursor (IL-20R-beta) (IL-20R2). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.022554 (rank : 110) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
NBN_MOUSE
|
||||||
| NC score | 0.022179 (rank : 111) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R207, O88981, Q3UY57, Q811I6, Q8CCY0, Q9R1X1 | Gene names | Nbn, Nbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1 homolog) (Cell cycle regulatory protein p95). | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.022164 (rank : 112) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.022059 (rank : 113) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.021934 (rank : 114) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
JOSD3_MOUSE
|
||||||
| NC score | 0.021870 (rank : 115) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D4V4, Q149X7, Q8C7I5, Q9CZG0, Q9D5U6 | Gene names | Josd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein JOSD3. | |||||
|
DJC12_HUMAN
|
||||||
| NC score | 0.021720 (rank : 116) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
BCAS3_MOUSE
|
||||||
| NC score | 0.021712 (rank : 117) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CCN5, Q8CC16, Q9EPX3 | Gene names | Bcas3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 3 homolog (K20D4). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.021428 (rank : 118) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
PCTL_HUMAN
|
||||||
| NC score | 0.020815 (rank : 119) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.020738 (rank : 120) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
KLC1_HUMAN
|
||||||
| NC score | 0.020210 (rank : 121) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.020177 (rank : 122) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.019669 (rank : 123) | θ value | 0.0736092 (rank : 38) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
CF081_MOUSE
|
||||||
| NC score | 0.019489 (rank : 124) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80X86, Q9D564, Q9DAN7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81 homolog. | |||||
|
PHC3_MOUSE
|
||||||
| NC score | 0.019469 (rank : 125) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
ZN292_HUMAN
|
||||||
| NC score | 0.019457 (rank : 126) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.019403 (rank : 127) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.019354 (rank : 128) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.019140 (rank : 129) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.018804 (rank : 130) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.018635 (rank : 131) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.018481 (rank : 132) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
XRN1_MOUSE
|
||||||
| NC score | 0.017954 (rank : 133) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97789, O35651, P97790, Q3TE44, Q3TRE9, Q3UQL5 | Gene names | Xrn1, Dhm2, Exo, Sep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (mXRN1) (Strand-exchange protein 1 homolog) (Protein Dhm2). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.017919 (rank : 134) | θ value | 0.0330416 (rank : 31) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
ESPL1_HUMAN
|
||||||
| NC score | 0.017857 (rank : 135) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14674 | Gene names | ESPL1, ESP1, KIAA0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.017090 (rank : 136) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.016482 (rank : 137) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.016327 (rank : 138) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.015620 (rank : 139) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.015571 (rank : 140) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
TR240_HUMAN
|
||||||
| NC score | 0.015145 (rank : 141) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHV7, O60334 | Gene names | THRAP1, ARC250, KIAA0593, TRAP240 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor-associated protein complex 240 kDa component (Trap240) (Thyroid hormone receptor-associated protein 1) (Vitamin D3 receptor-interacting protein complex component DRIP250) (DRIP 250) (Activator-recruited cofactor 250 kDa component) (ARC250). | |||||
|
K1024_MOUSE
|
||||||
| NC score | 0.015111 (rank : 142) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3V7, Q69ZS9 | Gene names | Kiaa1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024 (Protein DD1). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.015010 (rank : 143) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.014944 (rank : 144) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
K1024_HUMAN
|
||||||
| NC score | 0.014552 (rank : 145) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPX6 | Gene names | KIAA1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024. | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.013814 (rank : 146) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.013804 (rank : 147) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
NPHP3_HUMAN
|
||||||
| NC score | 0.013006 (rank : 148) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
CEBPB_HUMAN
|
||||||
| NC score | 0.012824 (rank : 149) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
NEBU_HUMAN
|
||||||
| NC score | 0.012306 (rank : 150) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |

|
|||||
| Description | Nebulin. | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.012237 (rank : 151) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
NONO_HUMAN
|
||||||
| NC score | 0.010947 (rank : 152) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |

|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
NONO_MOUSE
|
||||||
| NC score | 0.010795 (rank : 153) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
DYN1_MOUSE
|
||||||
| NC score | 0.009937 (rank : 154) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.009852 (rank : 155) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ANGP2_HUMAN
|
||||||
| NC score | 0.008306 (rank : 156) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15123, Q9NRR7, Q9P2Y7 | Gene names | ANGPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
RIT1_HUMAN
|
||||||
| NC score | 0.006142 (rank : 157) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92963, O00646, O00720 | Gene names | RIT1, RIBB, RIT, ROC1 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein Rit1 (Ras-like protein expressed in many tissues) (Ras-like without CAAX protein 1). | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.004976 (rank : 158) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
RIT1_MOUSE
|
||||||
| NC score | 0.003859 (rank : 159) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70426 | Gene names | Rit1, Rit | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein Rit1 (Ras-like protein expressed in many tissues) (Ras-like without CAAX protein 1). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.003390 (rank : 160) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
ZN688_HUMAN
|
||||||
| NC score | -0.002881 (rank : 161) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
ZN406_MOUSE
|
||||||
| NC score | -0.003089 (rank : 162) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TS63 | Gene names | Znf406, Zfp406 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406. | |||||
|
O5AT1_HUMAN
|
||||||
| NC score | -0.003266 (rank : 163) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHC5 | Gene names | OR5AT1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 5AT1. | |||||