Please be patient as the page loads
|
COPG2_MOUSE
|
||||||
| SwissProt Accessions | Q9QXK3 | Gene names | Copg2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
COPG2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999445 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBF2 | Gene names | COPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXK3 | Gene names | Copg2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.993190 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
AP4B1_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 4) | NC score | 0.444515 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WV76 | Gene names | Ap4b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta-1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP1B1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 5) | NC score | 0.416543 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP1B1_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 6) | NC score | 0.415731 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP2B1_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 7) | NC score | 0.412281 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 8) | NC score | 0.412292 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP4B1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 9) | NC score | 0.390242 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6B7 | Gene names | AP4B1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta 1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 10) | NC score | 0.254828 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 11) | NC score | 0.255492 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 12) | NC score | 0.256631 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 13) | NC score | 0.248861 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 14) | NC score | 0.224787 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
COPB_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.231935 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.233638 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP4E1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.188016 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
COPB_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.230511 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
AP4E1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.186652 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP1G2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.181435 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.179627 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
TPSN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.031493 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R233, O70317, Q91YE0, Q91YE1, Q9QWT7, Q9Z1W3 | Gene names | Tapbp, Tapa | |||
|
Domain Architecture |

|
|||||
| Description | Tapasin precursor (TPSN) (TPN) (TAP-binding protein) (TAP-associated protein). | |||||
|
DGLA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.019525 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6WQJ1, Q6ZQ76 | Gene names | Nsddr, Kiaa0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
IF4G2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.025863 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62448, P97867, Q921U3 | Gene names | Eif4g2, Nat1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Novel APOBEC-1 target 1) (Translation repressor NAT1). | |||||
|
CAND2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.027134 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75155 | Gene names | CAND2, KIAA0667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2). | |||||
|
CAND2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.026438 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
IF4G2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.023820 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78344, O60877, P78404, Q53EU1, Q58EZ2, Q8NI71, Q96C16 | Gene names | EIF4G2, DAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Death-associated protein 5) (DAP-5). | |||||
|
CASPA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.017836 (rank : 34) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |
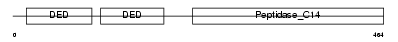
|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
FYB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.008589 (rank : 36) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.005166 (rank : 37) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.003952 (rank : 38) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
POPD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.009171 (rank : 35) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBU9, Q86UE7 | Gene names | POPDC2, POP2 | |||
|
Domain Architecture |

|
|||||
| Description | Popeye domain-containing protein 2 (Popeye protein 2). | |||||
|
AP2A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.124713 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.122486 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.133282 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.133078 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.204138 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.204925 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
COPG2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXK3 | Gene names | Copg2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG2_HUMAN
|
||||||
| NC score | 0.999445 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBF2 | Gene names | COPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG_HUMAN
|
||||||
| NC score | 0.993190 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
AP4B1_MOUSE
|
||||||
| NC score | 0.444515 (rank : 4) | θ value | 2.69671e-12 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WV76 | Gene names | Ap4b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta-1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP1B1_HUMAN
|
||||||
| NC score | 0.416543 (rank : 5) | θ value | 6.64225e-11 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP1B1_MOUSE
|
||||||
| NC score | 0.415731 (rank : 6) | θ value | 6.64225e-11 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP2B1_MOUSE
|
||||||
| NC score | 0.412292 (rank : 7) | θ value | 1.133e-10 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_HUMAN
|
||||||
| NC score | 0.412281 (rank : 8) | θ value | 1.133e-10 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP4B1_HUMAN
|
||||||
| NC score | 0.390242 (rank : 9) | θ value | 1.53129e-07 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6B7 | Gene names | AP4B1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta 1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.256631 (rank : 10) | θ value | 0.00228821 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.255492 (rank : 11) | θ value | 1.87187e-05 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.254828 (rank : 12) | θ value | 1.87187e-05 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.248861 (rank : 13) | θ value | 0.0148317 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.233638 (rank : 14) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
COPB_HUMAN
|
||||||
| NC score | 0.231935 (rank : 15) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPB_MOUSE
|
||||||
| NC score | 0.230511 (rank : 16) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.224787 (rank : 17) | θ value | 0.0148317 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.204925 (rank : 18) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.204138 (rank : 19) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
AP4E1_MOUSE
|
||||||
| NC score | 0.188016 (rank : 20) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_HUMAN
|
||||||
| NC score | 0.186652 (rank : 21) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP1G2_HUMAN
|
||||||
| NC score | 0.181435 (rank : 22) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.179627 (rank : 23) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP2A2_HUMAN
|
||||||
| NC score | 0.133282 (rank : 24) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| NC score | 0.133078 (rank : 25) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP2A1_HUMAN
|
||||||
| NC score | 0.124713 (rank : 26) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| NC score | 0.122486 (rank : 27) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
TPSN_MOUSE
|
||||||
| NC score | 0.031493 (rank : 28) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R233, O70317, Q91YE0, Q91YE1, Q9QWT7, Q9Z1W3 | Gene names | Tapbp, Tapa | |||
|
Domain Architecture |

|
|||||
| Description | Tapasin precursor (TPSN) (TPN) (TAP-binding protein) (TAP-associated protein). | |||||
|
CAND2_HUMAN
|
||||||
| NC score | 0.027134 (rank : 29) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75155 | Gene names | CAND2, KIAA0667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2). | |||||
|
CAND2_MOUSE
|
||||||
| NC score | 0.026438 (rank : 30) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
IF4G2_MOUSE
|
||||||
| NC score | 0.025863 (rank : 31) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62448, P97867, Q921U3 | Gene names | Eif4g2, Nat1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Novel APOBEC-1 target 1) (Translation repressor NAT1). | |||||
|
IF4G2_HUMAN
|
||||||
| NC score | 0.023820 (rank : 32) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78344, O60877, P78404, Q53EU1, Q58EZ2, Q8NI71, Q96C16 | Gene names | EIF4G2, DAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Death-associated protein 5) (DAP-5). | |||||
|
DGLA_MOUSE
|
||||||
| NC score | 0.019525 (rank : 33) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6WQJ1, Q6ZQ76 | Gene names | Nsddr, Kiaa0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
CASPA_HUMAN
|
||||||
| NC score | 0.017836 (rank : 34) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
POPD2_HUMAN
|
||||||
| NC score | 0.009171 (rank : 35) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBU9, Q86UE7 | Gene names | POPDC2, POP2 | |||
|
Domain Architecture |

|
|||||
| Description | Popeye domain-containing protein 2 (Popeye protein 2). | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.008589 (rank : 36) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.005166 (rank : 37) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.003952 (rank : 38) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||