Please be patient as the page loads
|
XRN1_MOUSE
|
||||||
| SwissProt Accessions | P97789, O35651, P97790, Q3TE44, Q3TRE9, Q3UQL5 | Gene names | Xrn1, Dhm2, Exo, Sep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (mXRN1) (Strand-exchange protein 1 homolog) (Protein Dhm2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
XRN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990918 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZH2, Q4G0S3, Q68D88, Q6AI24, Q6MZS8, Q86WS7, Q8N8U4, Q9UF39 | Gene names | XRN1, SEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (Strand-exchange protein 1 homolog). | |||||
|
XRN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P97789, O35651, P97790, Q3TE44, Q3TRE9, Q3UQL5 | Gene names | Xrn1, Dhm2, Exo, Sep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (mXRN1) (Strand-exchange protein 1 homolog) (Protein Dhm2). | |||||
|
XRN2_HUMAN
|
||||||
| θ value | 2.5521e-87 (rank : 3) | NC score | 0.839006 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0D6, Q3L8N4, Q6KGZ9, Q9BQL1, Q9NTW0, Q9NXS6, Q9UL53 | Gene names | XRN2 | |||
|
Domain Architecture |
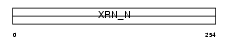
|
|||||
| Description | 5'-3' exoribonuclease 2 (EC 3.1.13.-) (DHM1-like protein) (DHP protein). | |||||
|
XRN2_MOUSE
|
||||||
| θ value | 2.5521e-87 (rank : 4) | NC score | 0.838761 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR1, Q3TI26, Q3TKF2, Q3UZT9, Q61489, Q99KS7 | Gene names | Xrn2, Dhm1 | |||
|
Domain Architecture |
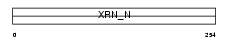
|
|||||
| Description | 5'-3' exoribonuclease 2 (EC 3.1.13.-) (Protein Dhm1). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 5) | NC score | 0.039568 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 6) | NC score | 0.033143 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TMC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.026882 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
FOS_HUMAN
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.040342 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
REXO4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.037008 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZR2, Q5T8S4, Q5T8S5, Q5T8S6, Q9GZW3 | Gene names | REXO4, PMC2, XPMC2H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (hPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
FOS_MOUSE
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.039149 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
IMP4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.057647 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHZ7, Q3U8H6, Q8CI42, Q9CWN6, Q9CYT5 | Gene names | Imp4, D1Wsu40e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein IMP4 (U3 snoRNP protein IMP4). | |||||
|
CP053_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.046920 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99L02 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf53 homolog. | |||||
|
JHD3D_MOUSE
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.018588 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.022723 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
DZIP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.021903 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
LAP2A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.025812 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
SYNG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.026365 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.009999 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EURL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.032083 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYK6, Q9H8C6 | Gene names | EURL, C21orf7, C21orf91, YG81 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EURL homolog. | |||||
|
GAB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.020092 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.013790 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SDCG1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.026104 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.020921 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
CH3L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.022283 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36222, P30923, Q8IVA4, Q96HI7 | Gene names | CHI3L1 | |||
|
Domain Architecture |
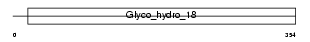
|
|||||
| Description | Chitinase-3-like protein 1 precursor (Cartilage glycoprotein 39) (GP- 39) (39 kDa synovial protein) (HCgp-39) (YKL-40). | |||||
|
CPNE8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.014873 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YQ8, Q2TB41 | Gene names | CPNE8 | |||
|
Domain Architecture |
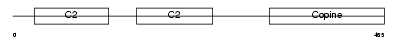
|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
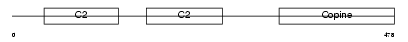
CPNE8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.014921 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DC53, Q9CUZ8 | Gene names | Cpne8 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.007692 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.015267 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.011855 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.009285 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.006963 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.005804 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
PRGC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.020946 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.007419 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
TULP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.018209 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.009907 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
UBXD8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.021447 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96CS3, O94963, Q8IUF2, Q9BRP2, Q9BVM7 | Gene names | UBXD8, ETEA, KIAA0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8 (Protein ETEA). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.009829 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
CDC20_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.007345 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12834, Q9BW56, Q9UQI9 | Gene names | CDC20 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 20 homolog (p55CDC). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.003564 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
FIBA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.012531 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02671, Q9BX62, Q9UCH2 | Gene names | FGA | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen alpha chain precursor [Contains: Fibrinopeptide A]. | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.012023 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TOP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.015617 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11387, O43256, Q12855, Q12856, Q15610, Q5TFY3, Q9UJN0 | Gene names | TOP1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.017954 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
LYAR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.019230 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NX58, Q6FI78, Q9NYS1 | Gene names | LYAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.006338 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
UDB11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.009183 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75310, Q3KNV9 | Gene names | UGT2B11 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 2B11 precursor (EC 2.4.1.17) (UDPGT). | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.013597 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
CEBPZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.013767 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03701, Q8NE75 | Gene names | CEBPZ, CBF2 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein zeta (CCAAT-box-binding transcription factor) (CCAAT-binding factor) (CBF). | |||||
|
CO4B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.007615 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01029, Q31201, Q61372, Q61859, Q62353 | Gene names | C4b, C4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C4-B precursor [Contains: Complement C4 beta chain; Complement C4 alpha chain; C4a anaphylatoxin; Complement C4 gamma chain]. | |||||
|
CP053_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.032412 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BTK6 | Gene names | C16orf53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf53. | |||||
|
DCC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.003467 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.012879 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
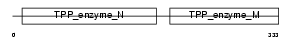
HPCL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.024652 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJ83, Q9BV42, Q9P0A2 | Gene names | HPCL, HPCL2, PHYH2 | |||
|
Domain Architecture |
|
|||||
| Description | 2-hydroxyphytanoyl-CoA lyase (EC 4.1.-.-) (2-HPCL). | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.013420 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.009287 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
TUB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.014240 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50607, O00293 | Gene names | TUB | |||
|
Domain Architecture |

|
|||||
| Description | Tubby protein homolog. | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.010486 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CELR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.004717 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
DLX5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.002694 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
GP1BA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.004679 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
K1383_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.015188 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2G4, Q58EZ9, Q5VV83 | Gene names | KIAA1383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1383. | |||||
|
NALD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.006503 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3Q0 | Gene names | NAALAD2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase 2 (EC 3.4.17.21) (N- acetylated-alpha-linked acidic dipeptidase II) (NAALADase II). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.010340 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
XRN1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P97789, O35651, P97790, Q3TE44, Q3TRE9, Q3UQL5 | Gene names | Xrn1, Dhm2, Exo, Sep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (mXRN1) (Strand-exchange protein 1 homolog) (Protein Dhm2). | |||||
|
XRN1_HUMAN
|
||||||
| NC score | 0.990918 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZH2, Q4G0S3, Q68D88, Q6AI24, Q6MZS8, Q86WS7, Q8N8U4, Q9UF39 | Gene names | XRN1, SEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (Strand-exchange protein 1 homolog). | |||||
|
XRN2_HUMAN
|
||||||
| NC score | 0.839006 (rank : 3) | θ value | 2.5521e-87 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0D6, Q3L8N4, Q6KGZ9, Q9BQL1, Q9NTW0, Q9NXS6, Q9UL53 | Gene names | XRN2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-3' exoribonuclease 2 (EC 3.1.13.-) (DHM1-like protein) (DHP protein). | |||||
|
XRN2_MOUSE
|
||||||
| NC score | 0.838761 (rank : 4) | θ value | 2.5521e-87 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR1, Q3TI26, Q3TKF2, Q3UZT9, Q61489, Q99KS7 | Gene names | Xrn2, Dhm1 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-3' exoribonuclease 2 (EC 3.1.13.-) (Protein Dhm1). | |||||
|
IMP4_MOUSE
|
||||||
| NC score | 0.057647 (rank : 5) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHZ7, Q3U8H6, Q8CI42, Q9CWN6, Q9CYT5 | Gene names | Imp4, D1Wsu40e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein IMP4 (U3 snoRNP protein IMP4). | |||||
|
CP053_MOUSE
|
||||||
| NC score | 0.046920 (rank : 6) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99L02 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf53 homolog. | |||||
|
FOS_HUMAN
|
||||||
| NC score | 0.040342 (rank : 7) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.039568 (rank : 8) | θ value | 0.21417 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
FOS_MOUSE
|
||||||
| NC score | 0.039149 (rank : 9) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
REXO4_HUMAN
|
||||||
| NC score | 0.037008 (rank : 10) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZR2, Q5T8S4, Q5T8S5, Q5T8S6, Q9GZW3 | Gene names | REXO4, PMC2, XPMC2H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (hPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.033143 (rank : 11) | θ value | 0.279714 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CP053_HUMAN
|
||||||
| NC score | 0.032412 (rank : 12) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BTK6 | Gene names | C16orf53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf53. | |||||
|
EURL_HUMAN
|
||||||
| NC score | 0.032083 (rank : 13) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYK6, Q9H8C6 | Gene names | EURL, C21orf7, C21orf91, YG81 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EURL homolog. | |||||
|
TMC2_HUMAN
|
||||||
| NC score | 0.026882 (rank : 14) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
SYNG_HUMAN
|
||||||
| NC score | 0.026365 (rank : 15) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
SDCG1_MOUSE
|
||||||
| NC score | 0.026104 (rank : 16) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
LAP2A_MOUSE
|
||||||
| NC score | 0.025812 (rank : 17) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
HPCL_HUMAN
|
||||||
| NC score | 0.024652 (rank : 18) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJ83, Q9BV42, Q9P0A2 | Gene names | HPCL, HPCL2, PHYH2 | |||
|
Domain Architecture |

|
|||||
| Description | 2-hydroxyphytanoyl-CoA lyase (EC 4.1.-.-) (2-HPCL). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.022723 (rank : 19) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CH3L1_HUMAN
|
||||||
| NC score | 0.022283 (rank : 20) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36222, P30923, Q8IVA4, Q96HI7 | Gene names | CHI3L1 | |||
|
Domain Architecture |
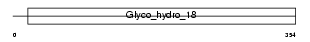
|
|||||
| Description | Chitinase-3-like protein 1 precursor (Cartilage glycoprotein 39) (GP- 39) (39 kDa synovial protein) (HCgp-39) (YKL-40). | |||||
|
DZIP1_HUMAN
|
||||||
| NC score | 0.021903 (rank : 21) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
UBXD8_HUMAN
|
||||||
| NC score | 0.021447 (rank : 22) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96CS3, O94963, Q8IUF2, Q9BRP2, Q9BVM7 | Gene names | UBXD8, ETEA, KIAA0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8 (Protein ETEA). | |||||
|
PRGC2_HUMAN
|
||||||
| NC score | 0.020946 (rank : 23) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.020921 (rank : 24) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
GAB2_HUMAN
|
||||||
| NC score | 0.020092 (rank : 25) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
LYAR_HUMAN
|
||||||
| NC score | 0.019230 (rank : 26) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NX58, Q6FI78, Q9NYS1 | Gene names | LYAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
JHD3D_MOUSE
|
||||||
| NC score | 0.018588 (rank : 27) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
TULP1_HUMAN
|
||||||
| NC score | 0.018209 (rank : 28) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.017954 (rank : 29) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
TOP1_HUMAN
|
||||||
| NC score | 0.015617 (rank : 30) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11387, O43256, Q12855, Q12856, Q15610, Q5TFY3, Q9UJN0 | Gene names | TOP1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.015267 (rank : 31) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
K1383_HUMAN
|
||||||
| NC score | 0.015188 (rank : 32) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2G4, Q58EZ9, Q5VV83 | Gene names | KIAA1383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1383. | |||||
|
CPNE8_MOUSE
|
||||||
| NC score | 0.014921 (rank : 33) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DC53, Q9CUZ8 | Gene names | Cpne8 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
CPNE8_HUMAN
|
||||||
| NC score | 0.014873 (rank : 34) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YQ8, Q2TB41 | Gene names | CPNE8 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
TUB_HUMAN
|
||||||
| NC score | 0.014240 (rank : 35) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50607, O00293 | Gene names | TUB | |||
|
Domain Architecture |

|
|||||
| Description | Tubby protein homolog. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.013790 (rank : 36) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CEBPZ_HUMAN
|
||||||
| NC score | 0.013767 (rank : 37) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03701, Q8NE75 | Gene names | CEBPZ, CBF2 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein zeta (CCAAT-box-binding transcription factor) (CCAAT-binding factor) (CBF). | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.013597 (rank : 38) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.013420 (rank : 39) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.012879 (rank : 40) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
FIBA_HUMAN
|
||||||
| NC score | 0.012531 (rank : 41) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02671, Q9BX62, Q9UCH2 | Gene names | FGA | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen alpha chain precursor [Contains: Fibrinopeptide A]. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.012023 (rank : 42) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.011855 (rank : 43) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.010486 (rank : 44) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.010340 (rank : 45) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.009999 (rank : 46) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.009907 (rank : 47) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.009829 (rank : 48) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.009287 (rank : 49) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.009285 (rank : 50) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
UDB11_HUMAN
|
||||||
| NC score | 0.009183 (rank : 51) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75310, Q3KNV9 | Gene names | UGT2B11 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 2B11 precursor (EC 2.4.1.17) (UDPGT). | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.007692 (rank : 52) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
CO4B_MOUSE
|
||||||
| NC score | 0.007615 (rank : 53) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01029, Q31201, Q61372, Q61859, Q62353 | Gene names | C4b, C4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C4-B precursor [Contains: Complement C4 beta chain; Complement C4 alpha chain; C4a anaphylatoxin; Complement C4 gamma chain]. | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.007419 (rank : 54) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
CDC20_HUMAN
|
||||||
| NC score | 0.007345 (rank : 55) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12834, Q9BW56, Q9UQI9 | Gene names | CDC20 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 20 homolog (p55CDC). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.006963 (rank : 56) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NALD2_HUMAN
|
||||||
| NC score | 0.006503 (rank : 57) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3Q0 | Gene names | NAALAD2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase 2 (EC 3.4.17.21) (N- acetylated-alpha-linked acidic dipeptidase II) (NAALADase II). | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.006338 (rank : 58) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.005804 (rank : 59) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CELR1_HUMAN
|
||||||
| NC score | 0.004717 (rank : 60) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
GP1BA_HUMAN
|
||||||
| NC score | 0.004679 (rank : 61) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.003564 (rank : 62) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
DCC_HUMAN
|
||||||
| NC score | 0.003467 (rank : 63) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
DLX5_MOUSE
|
||||||
| NC score | 0.002694 (rank : 64) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||