Please be patient as the page loads
|
STAP2_MOUSE
|
||||||
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STAP2_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
STAP2_HUMAN
|
||||||
| θ value | 1.63136e-134 (rank : 2) | NC score | 0.977047 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGK3, Q9NXI2 | Gene names | STAP2, BKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2) (Breast tumor kinase substrate) (BRK substrate). | |||||
|
STAP1_HUMAN
|
||||||
| θ value | 1.28434e-38 (rank : 3) | NC score | 0.836040 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULZ2 | Gene names | STAP1, BRDG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 1 (STAP-1) (Stem cell adaptor protein 1) (BCR downstream signaling protein 1) (Docking protein BRDG1). | |||||
|
STAP1_MOUSE
|
||||||
| θ value | 1.08726e-37 (rank : 4) | NC score | 0.833047 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JM90, Q3TRM1, Q6PES6 | Gene names | Stap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 1 (STAP-1) (Stem cell adaptor protein 1). | |||||
|
PLCG2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 5) | NC score | 0.097072 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16885, Q969T5 | Gene names | PLCG2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-2) (Phospholipase C-gamma-2) (PLC-IV). | |||||
|
BLK_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 6) | NC score | 0.041593 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16277 | Gene names | Blk | |||
|
Domain Architecture |
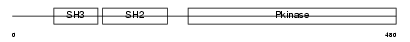
|
|||||
| Description | Tyrosine-protein kinase BLK (EC 2.7.10.2) (B lymphocyte kinase) (p55- BLK). | |||||
|
SRMS_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.041207 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62270, Q62360, Q923M5 | Gene names | Srms, Srm | |||
|
Domain Architecture |
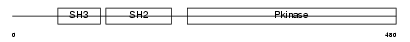
|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2) (PTK70). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.044102 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
SRMS_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.042208 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H3Y6 | Gene names | SRMS, C20orf148 | |||
|
Domain Architecture |
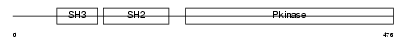
|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2). | |||||
|
BLK_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.038792 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P51451, Q16291 | Gene names | BLK | |||
|
Domain Architecture |
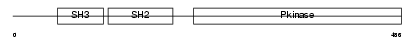
|
|||||
| Description | Tyrosine-protein kinase BLK (EC 2.7.10.2) (B lymphocyte kinase) (p55- BLK). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.032202 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
AKA11_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.062531 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKA4, O75124, Q9NUK7 | Gene names | AKAP11, AKAP220, KIAA0629 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 11 (Protein kinase A-anchoring protein 11) (PRKA11) (A kinase anchor protein 220 kDa) (AKAP 220) (hAKAP220). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.035270 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
ITK_MOUSE
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.031681 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 985 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q03526 | Gene names | Itk, Emt, Tlk, Tsk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ITK/TSK (EC 2.7.10.2) (T-cell-specific kinase) (IL-2-inducible T-cell kinase) (Kinase EMT) (Kinase TLK). | |||||
|
FRK_HUMAN
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.032472 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42685, Q13128 | Gene names | FRK | |||
|
Domain Architecture |
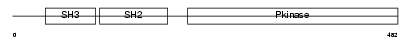
|
|||||
| Description | Tyrosine-protein kinase FRK (EC 2.7.10.2) (Nuclear tyrosine protein kinase RAK). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.037260 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.023965 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.036789 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
LCK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.034000 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06239, P07100, Q12850, Q13152, Q5TDH8, Q5TDH9, Q96DW4, Q9NYT8 | Gene names | LCK | |||
|
Domain Architecture |
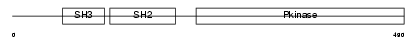
|
|||||
| Description | Proto-oncogene tyrosine-protein kinase LCK (EC 2.7.10.2) (p56-LCK) (Lymphocyte cell-specific protein-tyrosine kinase) (LSK) (T cell- specific protein-tyrosine kinase). | |||||
|
BLNK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.044192 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
ERBB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.031919 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.020535 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
IDD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.025145 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |
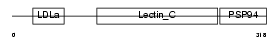
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
LCK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.033568 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06240, Q61794, Q61795, Q62320, Q91X65 | Gene names | Lck, Lsk-t | |||
|
Domain Architecture |
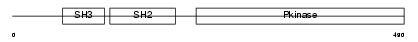
|
|||||
| Description | Proto-oncogene tyrosine-protein kinase LCK (EC 2.7.10.2) (p56-LCK) (Lymphocyte cell-specific protein-tyrosine kinase) (LSK). | |||||
|
SON_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.019180 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.022270 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
FARP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.022052 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.019900 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SACS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.031382 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
CBL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.018346 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
FOXO4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.009549 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |
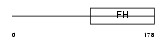
|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
NCK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.043572 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P16333 | Gene names | NCK1, NCK | |||
|
Domain Architecture |
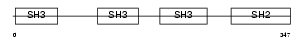
|
|||||
| Description | Cytoplasmic protein NCK1 (NCK adaptor protein 1) (SH2/SH3 adaptor protein NCK-alpha). | |||||
|
SACS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.030324 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.027305 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
ZFYV9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.012000 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
K0174_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.015961 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CX00, Q80U68 | Gene names | Kiaa0174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0174. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.009190 (rank : 44) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.006068 (rank : 45) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SSH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.011077 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.010581 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
TF3C3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.017109 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
SLAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.053013 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13239, Q9UMQ8 | Gene names | SLA, SLAP, SLAP1 | |||
|
Domain Architecture |
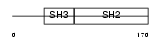
|
|||||
| Description | SRC-like-adapter (Src-like-adapter protein 1) (hSLAP). | |||||
|
SLAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.053659 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60898, Q8C9Q8, Q8CAT0, Q8CBE9, Q8QZX8 | Gene names | Sla, Slap, Slap1 | |||
|
Domain Architecture |
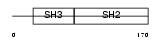
|
|||||
| Description | SRC-like-adapter (Src-like-adapter protein 1) (mSLAP). | |||||
|
SLAP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.055427 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H6Q3, Q5TH27, Q5TH28, Q8WY18, Q96QI4, Q9H135 | Gene names | SLA2, C20orf156, SLAP2 | |||
|
Domain Architecture |
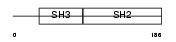
|
|||||
| Description | Src-like-adapter 2 (Src-like adapter protein 2) (SLAP-2) (Modulator of antigen receptor signaling) (MARS). | |||||
|
SLAP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.058417 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4L0, Q3UU74, Q8C0K2, Q8VI42, Q9D1Z9 | Gene names | Sla2 | |||
|
Domain Architecture |
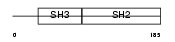
|
|||||
| Description | Src-like-adapter 2 (Src-like adapter protein 2) (SLAP-2). | |||||
|
STAP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
STAP2_HUMAN
|
||||||
| NC score | 0.977047 (rank : 2) | θ value | 1.63136e-134 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGK3, Q9NXI2 | Gene names | STAP2, BKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2) (Breast tumor kinase substrate) (BRK substrate). | |||||
|
STAP1_HUMAN
|
||||||
| NC score | 0.836040 (rank : 3) | θ value | 1.28434e-38 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULZ2 | Gene names | STAP1, BRDG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 1 (STAP-1) (Stem cell adaptor protein 1) (BCR downstream signaling protein 1) (Docking protein BRDG1). | |||||
|
STAP1_MOUSE
|
||||||
| NC score | 0.833047 (rank : 4) | θ value | 1.08726e-37 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JM90, Q3TRM1, Q6PES6 | Gene names | Stap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 1 (STAP-1) (Stem cell adaptor protein 1). | |||||
|
PLCG2_HUMAN
|
||||||
| NC score | 0.097072 (rank : 5) | θ value | 0.00102713 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16885, Q969T5 | Gene names | PLCG2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-2) (Phospholipase C-gamma-2) (PLC-IV). | |||||
|
AKA11_HUMAN
|
||||||
| NC score | 0.062531 (rank : 6) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKA4, O75124, Q9NUK7 | Gene names | AKAP11, AKAP220, KIAA0629 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 11 (Protein kinase A-anchoring protein 11) (PRKA11) (A kinase anchor protein 220 kDa) (AKAP 220) (hAKAP220). | |||||
|
SLAP2_MOUSE
|
||||||
| NC score | 0.058417 (rank : 7) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4L0, Q3UU74, Q8C0K2, Q8VI42, Q9D1Z9 | Gene names | Sla2 | |||
|
Domain Architecture |

|
|||||
| Description | Src-like-adapter 2 (Src-like adapter protein 2) (SLAP-2). | |||||
|
SLAP2_HUMAN
|
||||||
| NC score | 0.055427 (rank : 8) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H6Q3, Q5TH27, Q5TH28, Q8WY18, Q96QI4, Q9H135 | Gene names | SLA2, C20orf156, SLAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Src-like-adapter 2 (Src-like adapter protein 2) (SLAP-2) (Modulator of antigen receptor signaling) (MARS). | |||||
|
SLAP1_MOUSE
|
||||||
| NC score | 0.053659 (rank : 9) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60898, Q8C9Q8, Q8CAT0, Q8CBE9, Q8QZX8 | Gene names | Sla, Slap, Slap1 | |||
|
Domain Architecture |

|
|||||
| Description | SRC-like-adapter (Src-like-adapter protein 1) (mSLAP). | |||||
|
SLAP1_HUMAN
|
||||||
| NC score | 0.053013 (rank : 10) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13239, Q9UMQ8 | Gene names | SLA, SLAP, SLAP1 | |||
|
Domain Architecture |

|
|||||
| Description | SRC-like-adapter (Src-like-adapter protein 1) (hSLAP). | |||||
|
BLNK_MOUSE
|
||||||
| NC score | 0.044192 (rank : 11) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.044102 (rank : 12) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
NCK1_HUMAN
|
||||||
| NC score | 0.043572 (rank : 13) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P16333 | Gene names | NCK1, NCK | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic protein NCK1 (NCK adaptor protein 1) (SH2/SH3 adaptor protein NCK-alpha). | |||||
|
SRMS_HUMAN
|
||||||
| NC score | 0.042208 (rank : 14) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H3Y6 | Gene names | SRMS, C20orf148 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2). | |||||
|
BLK_MOUSE
|
||||||
| NC score | 0.041593 (rank : 15) | θ value | 0.00509761 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16277 | Gene names | Blk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase BLK (EC 2.7.10.2) (B lymphocyte kinase) (p55- BLK). | |||||
|
SRMS_MOUSE
|
||||||
| NC score | 0.041207 (rank : 16) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62270, Q62360, Q923M5 | Gene names | Srms, Srm | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2) (PTK70). | |||||
|
BLK_HUMAN
|
||||||
| NC score | 0.038792 (rank : 17) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P51451, Q16291 | Gene names | BLK | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase BLK (EC 2.7.10.2) (B lymphocyte kinase) (p55- BLK). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.037260 (rank : 18) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.036789 (rank : 19) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.035270 (rank : 20) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
LCK_HUMAN
|
||||||
| NC score | 0.034000 (rank : 21) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06239, P07100, Q12850, Q13152, Q5TDH8, Q5TDH9, Q96DW4, Q9NYT8 | Gene names | LCK | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase LCK (EC 2.7.10.2) (p56-LCK) (Lymphocyte cell-specific protein-tyrosine kinase) (LSK) (T cell- specific protein-tyrosine kinase). | |||||
|
LCK_MOUSE
|
||||||
| NC score | 0.033568 (rank : 22) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06240, Q61794, Q61795, Q62320, Q91X65 | Gene names | Lck, Lsk-t | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase LCK (EC 2.7.10.2) (p56-LCK) (Lymphocyte cell-specific protein-tyrosine kinase) (LSK). | |||||
|
FRK_HUMAN
|
||||||
| NC score | 0.032472 (rank : 23) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42685, Q13128 | Gene names | FRK | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase FRK (EC 2.7.10.2) (Nuclear tyrosine protein kinase RAK). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.032202 (rank : 24) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ERBB2_MOUSE
|
||||||
| NC score | 0.031919 (rank : 25) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||
|
ITK_MOUSE
|
||||||
| NC score | 0.031681 (rank : 26) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 985 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q03526 | Gene names | Itk, Emt, Tlk, Tsk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ITK/TSK (EC 2.7.10.2) (T-cell-specific kinase) (IL-2-inducible T-cell kinase) (Kinase EMT) (Kinase TLK). | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.031382 (rank : 27) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.030324 (rank : 28) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.027305 (rank : 29) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
IDD_MOUSE
|
||||||
| NC score | 0.025145 (rank : 30) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.023965 (rank : 31) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.022270 (rank : 32) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.022052 (rank : 33) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.020535 (rank : 34) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.019900 (rank : 35) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.019180 (rank : 36) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CBL_HUMAN
|
||||||
| NC score | 0.018346 (rank : 37) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
TF3C3_HUMAN
|
||||||
| NC score | 0.017109 (rank : 38) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
K0174_MOUSE
|
||||||
| NC score | 0.015961 (rank : 39) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CX00, Q80U68 | Gene names | Kiaa0174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0174. | |||||
|
ZFYV9_HUMAN
|
||||||
| NC score | 0.012000 (rank : 40) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
SSH1_HUMAN
|
||||||
| NC score | 0.011077 (rank : 41) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.010581 (rank : 42) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
FOXO4_MOUSE
|
||||||
| NC score | 0.009549 (rank : 43) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.009190 (rank : 44) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.006068 (rank : 45) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||