Please be patient as the page loads
|
ODPX_HUMAN
|
||||||
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ODPX_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
ODPX_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992209 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
ODP2_HUMAN
|
||||||
| θ value | 1.8332e-77 (rank : 3) | NC score | 0.927740 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
ODB2_HUMAN
|
||||||
| θ value | 4.72714e-33 (rank : 4) | NC score | 0.807893 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |
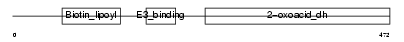
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_MOUSE
|
||||||
| θ value | 4.89182e-30 (rank : 5) | NC score | 0.800959 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |
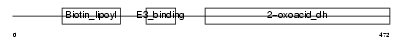
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODO2_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 6) | NC score | 0.757004 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODO2_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 7) | NC score | 0.750558 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
RFIP1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 8) | NC score | 0.104571 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
MCCA_MOUSE
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.088473 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
COA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.054680 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
FOG2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.021719 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WW38, Q9NPL7, Q9NPS4, Q9UNI5 | Gene names | ZFPM2, FOG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (FOG-2) (hFOG-2). | |||||
|
MCCA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.087384 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.015672 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
SYP2L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.024909 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
GRB7_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.015262 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03160 | Gene names | Grb7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth factor receptor-bound protein 7 (GRB7 adapter protein) (Epidermal growth factor receptor GRB-7). | |||||
|
FOG2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.018553 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CCH7, Q9Z0F2 | Gene names | Zfpm2, Fog2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (Friend of GATA-2) (FOG-2) (mFOG-2). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.010485 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.012420 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.016239 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.025422 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.025513 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.004457 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.007265 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PROX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.016203 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
ODPX_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
ODPX_MOUSE
|
||||||
| NC score | 0.992209 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
ODP2_HUMAN
|
||||||
| NC score | 0.927740 (rank : 3) | θ value | 1.8332e-77 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
ODB2_HUMAN
|
||||||
| NC score | 0.807893 (rank : 4) | θ value | 4.72714e-33 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_MOUSE
|
||||||
| NC score | 0.800959 (rank : 5) | θ value | 4.89182e-30 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODO2_HUMAN
|
||||||
| NC score | 0.757004 (rank : 6) | θ value | 5.59698e-26 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODO2_MOUSE
|
||||||
| NC score | 0.750558 (rank : 7) | θ value | 1.24688e-25 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
RFIP1_MOUSE
|
||||||
| NC score | 0.104571 (rank : 8) | θ value | 0.000786445 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
MCCA_MOUSE
|
||||||
| NC score | 0.088473 (rank : 9) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_HUMAN
|
||||||
| NC score | 0.087384 (rank : 10) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
COA1_HUMAN
|
||||||
| NC score | 0.054680 (rank : 11) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.025513 (rank : 12) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.025422 (rank : 13) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SYP2L_MOUSE
|
||||||
| NC score | 0.024909 (rank : 14) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
FOG2_HUMAN
|
||||||
| NC score | 0.021719 (rank : 15) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WW38, Q9NPL7, Q9NPS4, Q9UNI5 | Gene names | ZFPM2, FOG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (FOG-2) (hFOG-2). | |||||
|
FOG2_MOUSE
|
||||||
| NC score | 0.018553 (rank : 16) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CCH7, Q9Z0F2 | Gene names | Zfpm2, Fog2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (Friend of GATA-2) (FOG-2) (mFOG-2). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.016239 (rank : 17) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PROX1_MOUSE
|
||||||
| NC score | 0.016203 (rank : 18) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.015672 (rank : 19) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
GRB7_MOUSE
|
||||||
| NC score | 0.015262 (rank : 20) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03160 | Gene names | Grb7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth factor receptor-bound protein 7 (GRB7 adapter protein) (Epidermal growth factor receptor GRB-7). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.012420 (rank : 21) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.010485 (rank : 22) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.007265 (rank : 23) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.004457 (rank : 24) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||