Please be patient as the page loads
|
PIPNB_HUMAN
|
||||||
| SwissProt Accessions | P48739, Q8N5W1 | Gene names | PITPNB | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PIPNB_HUMAN
|
||||||
| θ value | 2.04462e-161 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48739, Q8N5W1 | Gene names | PITPNB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
PIPNB_MOUSE
|
||||||
| θ value | 6.1562e-158 (rank : 2) | NC score | 0.999516 (rank : 2) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P53811 | Gene names | Pitpnb | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
PIPNA_HUMAN
|
||||||
| θ value | 1.86652e-130 (rank : 3) | NC score | 0.998290 (rank : 3) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00169 | Gene names | PITPNA, PITPN | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNA_MOUSE
|
||||||
| θ value | 2.06368e-129 (rank : 4) | NC score | 0.998284 (rank : 4) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53810 | Gene names | Pitpna, Pitpn | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PITM2_MOUSE
|
||||||
| θ value | 3.84025e-59 (rank : 5) | NC score | 0.771167 (rank : 7) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
PITM1_MOUSE
|
||||||
| θ value | 1.11734e-58 (rank : 6) | NC score | 0.776567 (rank : 5) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
PITM1_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 7) | NC score | 0.770502 (rank : 8) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
PITM2_HUMAN
|
||||||
| θ value | 3.25095e-58 (rank : 8) | NC score | 0.775897 (rank : 6) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
ATS20_MOUSE
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.007009 (rank : 19) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59511 | Gene names | Adamts20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
TFAM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.020361 (rank : 17) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
MKL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.017908 (rank : 18) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
SIN3A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.024653 (rank : 16) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.024773 (rank : 15) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | -0.000419 (rank : 21) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | -0.000249 (rank : 20) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
DDHD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 16) | NC score | 0.129146 (rank : 12) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
DDHD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 17) | NC score | 0.135675 (rank : 11) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YA3, Q8VEF2 | Gene names | Ddhd1 | |||
|
Domain Architecture |
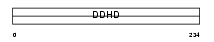
|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
PITM3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 18) | NC score | 0.438443 (rank : 10) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZ71, Q59GH9, Q9NPQ4 | Gene names | PITPNM3, NIR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PITM3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.441744 (rank : 9) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UHE1, Q3UH22, Q5RIT9 | Gene names | Pitpnm3, Nir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
S23IP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 20) | NC score | 0.111446 (rank : 13) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.109548 (rank : 14) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
PIPNB_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.04462e-161 (rank : 1) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48739, Q8N5W1 | Gene names | PITPNB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
PIPNB_MOUSE
|
||||||
| NC score | 0.999516 (rank : 2) | θ value | 6.1562e-158 (rank : 2) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P53811 | Gene names | Pitpnb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
PIPNA_HUMAN
|
||||||
| NC score | 0.998290 (rank : 3) | θ value | 1.86652e-130 (rank : 3) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00169 | Gene names | PITPNA, PITPN | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNA_MOUSE
|
||||||
| NC score | 0.998284 (rank : 4) | θ value | 2.06368e-129 (rank : 4) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53810 | Gene names | Pitpna, Pitpn | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PITM1_MOUSE
|
||||||
| NC score | 0.776567 (rank : 5) | θ value | 1.11734e-58 (rank : 6) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
PITM2_HUMAN
|
||||||
| NC score | 0.775897 (rank : 6) | θ value | 3.25095e-58 (rank : 8) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
PITM2_MOUSE
|
||||||
| NC score | 0.771167 (rank : 7) | θ value | 3.84025e-59 (rank : 5) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
PITM1_HUMAN
|
||||||
| NC score | 0.770502 (rank : 8) | θ value | 2.48917e-58 (rank : 7) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
PITM3_MOUSE
|
||||||
| NC score | 0.441744 (rank : 9) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UHE1, Q3UH22, Q5RIT9 | Gene names | Pitpnm3, Nir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PITM3_HUMAN
|
||||||
| NC score | 0.438443 (rank : 10) | θ value | θ > 10 (rank : 18) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZ71, Q59GH9, Q9NPQ4 | Gene names | PITPNM3, NIR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
DDHD1_MOUSE
|
||||||
| NC score | 0.135675 (rank : 11) | θ value | θ > 10 (rank : 17) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YA3, Q8VEF2 | Gene names | Ddhd1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
DDHD1_HUMAN
|
||||||
| NC score | 0.129146 (rank : 12) | θ value | θ > 10 (rank : 16) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
S23IP_HUMAN
|
||||||
| NC score | 0.111446 (rank : 13) | θ value | θ > 10 (rank : 20) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.109548 (rank : 14) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.024773 (rank : 15) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SIN3A_HUMAN
|
||||||
| NC score | 0.024653 (rank : 16) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
TFAM_HUMAN
|
||||||
| NC score | 0.020361 (rank : 17) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
MKL2_MOUSE
|
||||||
| NC score | 0.017908 (rank : 18) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
ATS20_MOUSE
|
||||||
| NC score | 0.007009 (rank : 19) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59511 | Gene names | Adamts20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | -0.000249 (rank : 20) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | -0.000419 (rank : 21) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||