Please be patient as the page loads
|
DEND_HUMAN
|
||||||
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DEND_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
DEND_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.951638 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80TS7 | Gene names | Ddn, Kiaa0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
DOK7_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 3) | NC score | 0.111622 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
ZN688_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 4) | NC score | 0.017471 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
PLCB2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 5) | NC score | 0.040014 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
SORC3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.042000 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 7) | NC score | 0.040868 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
KLC4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.044515 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.032704 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CT128_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.071056 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.052777 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PALB2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.059805 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
PP2CG_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.047764 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.058883 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.045093 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CXCC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.051062 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |
|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.032028 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.043629 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.057210 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.049456 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.036025 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.038890 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
BOP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.028729 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97452, Q3TK87, Q91X31 | Gene names | Bop1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BOP1 (Block of proliferation 1 protein). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.026098 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
BMP8B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.015025 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55105 | Gene names | Bmp8b | |||
|
Domain Architecture |
|
|||||
| Description | Bone morphogenetic protein 8B precursor (BMP-8B). | |||||
|
FHOD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.031818 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.016213 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
RPKL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.010767 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R2S1, Q8R1D4 | Gene names | Rps6kl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase-like 1 (EC 2.7.11.1). | |||||
|
SP6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.011705 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3SY56 | Gene names | SP6, KLF14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14). | |||||
|
SP6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.011694 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESX2, Q6VM22 | Gene names | Sp6, Epfn, Klf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14) (Epiprofin). | |||||
|
BPL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.036038 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50747, Q99451 | Gene names | HLCS | |||
|
Domain Architecture |

|
|||||
| Description | Biotin--protein ligase (EC 6.3.4.-) (Biotin apo-protein ligase) [Includes: Biotin--[methylmalonyl-CoA-carboxytransferase] ligase (EC 6.3.4.9); Biotin--[propionyl-CoA-carboxylase [ATP-hydrolyzing]] ligase (EC 6.3.4.10) (Holocarboxylase synthetase) (HCS); Biotin-- [methylcrotonoyl-CoA-carboxylase] ligase (EC 6.3.4.11); Biotin-- [acetyl-CoA-carboxylase] ligase (EC 6.3.4.15)]. | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.042687 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.029758 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
ASH2L_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.032536 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.021555 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.023331 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KLC4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.035845 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
LPIN3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.021199 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.061816 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.054157 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
ARMX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.026944 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
IP3KC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.023911 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DU7, Q9UE25, Q9Y475 | Gene names | ITPKC, IP3KC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (InsP 3-kinase C) (IP3K-C). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.034013 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
TFE2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.019739 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
WDR46_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.046254 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |
|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.037735 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
ATS12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.007759 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q811B3, Q8BK92, Q8BKY1 | Gene names | Adamts12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
CBX4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.021379 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.035099 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.029508 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MK15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.002219 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.006663 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.024307 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.017747 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.051246 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.033821 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.023085 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
HORN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.031020 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.020521 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
KCD17_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.014580 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N5Z5, O95517 | Gene names | KCTD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD17. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.029650 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.036752 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.036639 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.025378 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.007350 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SSDH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.009932 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWF0 | Gene names | Aldh5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
TM16H_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.021813 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PB70, Q69ZE4 | Gene names | Tmem16h, Kiaa1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
FBXW8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.015791 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BIA4, Q8BI62, Q8BI75, Q8BI76, Q8CID8, Q921Z1 | Gene names | Fbxw8 | |||
|
Domain Architecture |
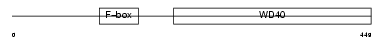
|
|||||
| Description | F-box/WD repeat protein 8 (F-box and WD-40 domain protein 8). | |||||
|
GMEB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.015161 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58929, Q6PCY0 | Gene names | Gmeb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucocorticoid modulatory element-binding protein 2 (GMEB-2). | |||||
|
K0310_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.037364 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
NEST_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.012736 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PITM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.011297 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
PRD15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.005702 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.036859 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SF04_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.030101 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
TBX21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.009696 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.018611 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CE016_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.016381 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.007504 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DOS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.037769 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N350, O43385 | Gene names | DOS, C19orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dos. | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.019656 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
K1949_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.025983 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.009499 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.025343 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PML_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.015523 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P29590, P29591, P29592, P29593, Q00755 | Gene names | PML, MYL, RNF71, TRIM19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable transcription factor PML (Tripartite motif-containing protein 19) (RING finger protein 71). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.018737 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.037419 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ZN263_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.003026 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14978, O43387, Q96H95 | Gene names | ZNF263, FPM315 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 263 (Zinc finger protein FPM315). | |||||
|
DOK7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.059245 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q18PE1, Q6P6A6, Q86XG5, Q8N2J3, Q8NBC1 | Gene names | DOK7, C4orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.050775 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
DEND_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
DEND_MOUSE
|
||||||
| NC score | 0.951638 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80TS7 | Gene names | Ddn, Kiaa0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
DOK7_MOUSE
|
||||||
| NC score | 0.111622 (rank : 3) | θ value | 0.00509761 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
CT128_HUMAN
|
||||||
| NC score | 0.071056 (rank : 4) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.061816 (rank : 5) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
PALB2_MOUSE
|
||||||
| NC score | 0.059805 (rank : 6) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
DOK7_HUMAN
|
||||||
| NC score | 0.059245 (rank : 7) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q18PE1, Q6P6A6, Q86XG5, Q8N2J3, Q8NBC1 | Gene names | DOK7, C4orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.058883 (rank : 8) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.057210 (rank : 9) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.054157 (rank : 10) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.052777 (rank : 11) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.051246 (rank : 12) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CXCC1_HUMAN
|
||||||
| NC score | 0.051062 (rank : 13) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.050775 (rank : 14) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.049456 (rank : 15) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
PP2CG_HUMAN
|
||||||
| NC score | 0.047764 (rank : 16) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
WDR46_MOUSE
|
||||||
| NC score | 0.046254 (rank : 17) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.045093 (rank : 18) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
KLC4_HUMAN
|
||||||
| NC score | 0.044515 (rank : 19) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.043629 (rank : 20) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.042687 (rank : 21) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
SORC3_HUMAN
|
||||||
| NC score | 0.042000 (rank : 22) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.040868 (rank : 23) | θ value | 0.0961366 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
PLCB2_HUMAN
|
||||||
| NC score | 0.040014 (rank : 24) | θ value | 0.0330416 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.038890 (rank : 25) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
DOS_HUMAN
|
||||||
| NC score | 0.037769 (rank : 26) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N350, O43385 | Gene names | DOS, C19orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dos. | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.037735 (rank : 27) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.037419 (rank : 28) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.037364 (rank : 29) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.036859 (rank : 30) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.036752 (rank : 31) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.036639 (rank : 32) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
BPL1_HUMAN
|
||||||
| NC score | 0.036038 (rank : 33) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50747, Q99451 | Gene names | HLCS | |||
|
Domain Architecture |

|
|||||
| Description | Biotin--protein ligase (EC 6.3.4.-) (Biotin apo-protein ligase) [Includes: Biotin--[methylmalonyl-CoA-carboxytransferase] ligase (EC 6.3.4.9); Biotin--[propionyl-CoA-carboxylase [ATP-hydrolyzing]] ligase (EC 6.3.4.10) (Holocarboxylase synthetase) (HCS); Biotin-- [methylcrotonoyl-CoA-carboxylase] ligase (EC 6.3.4.11); Biotin-- [acetyl-CoA-carboxylase] ligase (EC 6.3.4.15)]. | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.036025 (rank : 34) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
KLC4_MOUSE
|
||||||
| NC score | 0.035845 (rank : 35) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.035099 (rank : 36) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.034013 (rank : 37) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.033821 (rank : 38) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.032704 (rank : 39) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ASH2L_MOUSE
|
||||||
| NC score | 0.032536 (rank : 40) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.032028 (rank : 41) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
FHOD1_HUMAN
|
||||||
| NC score | 0.031818 (rank : 42) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.031020 (rank : 43) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
SF04_HUMAN
|
||||||
| NC score | 0.030101 (rank : 44) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.029758 (rank : 45) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.029650 (rank : 46) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.029508 (rank : 47) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
BOP1_MOUSE
|
||||||
| NC score | 0.028729 (rank : 48) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97452, Q3TK87, Q91X31 | Gene names | Bop1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BOP1 (Block of proliferation 1 protein). | |||||
|
ARMX2_HUMAN
|
||||||
| NC score | 0.026944 (rank : 49) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.026098 (rank : 50) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
K1949_MOUSE
|
||||||
| NC score | 0.025983 (rank : 51) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.025378 (rank : 52) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.025343 (rank : 53) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.024307 (rank : 54) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
IP3KC_HUMAN
|
||||||
| NC score | 0.023911 (rank : 55) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DU7, Q9UE25, Q9Y475 | Gene names | ITPKC, IP3KC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (InsP 3-kinase C) (IP3K-C). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.023331 (rank : 56) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.023085 (rank : 57) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
TM16H_MOUSE
|
||||||
| NC score | 0.021813 (rank : 58) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PB70, Q69ZE4 | Gene names | Tmem16h, Kiaa1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.021555 (rank : 59) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CBX4_MOUSE
|
||||||
| NC score | 0.021379 (rank : 60) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
LPIN3_HUMAN
|
||||||
| NC score | 0.021199 (rank : 61) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.020521 (rank : 62) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
TFE2_MOUSE
|
||||||
| NC score | 0.019739 (rank : 63) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.019656 (rank : 64) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.018737 (rank : 65) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.018611 (rank : 66) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.017747 (rank : 67) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ZN688_HUMAN
|
||||||
| NC score | 0.017471 (rank : 68) | θ value | 0.00665767 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
CE016_HUMAN
|
||||||
| NC score | 0.016381 (rank : 69) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.016213 (rank : 70) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
FBXW8_MOUSE
|
||||||
| NC score | 0.015791 (rank : 71) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BIA4, Q8BI62, Q8BI75, Q8BI76, Q8CID8, Q921Z1 | Gene names | Fbxw8 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 8 (F-box and WD-40 domain protein 8). | |||||
|
PML_HUMAN
|
||||||
| NC score | 0.015523 (rank : 72) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P29590, P29591, P29592, P29593, Q00755 | Gene names | PML, MYL, RNF71, TRIM19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable transcription factor PML (Tripartite motif-containing protein 19) (RING finger protein 71). | |||||
|
GMEB2_MOUSE
|
||||||
| NC score | 0.015161 (rank : 73) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58929, Q6PCY0 | Gene names | Gmeb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucocorticoid modulatory element-binding protein 2 (GMEB-2). | |||||
|
BMP8B_MOUSE
|
||||||
| NC score | 0.015025 (rank : 74) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55105 | Gene names | Bmp8b | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 8B precursor (BMP-8B). | |||||
|
KCD17_HUMAN
|
||||||
| NC score | 0.014580 (rank : 75) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N5Z5, O95517 | Gene names | KCTD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD17. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.012736 (rank : 76) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
SP6_HUMAN
|
||||||
| NC score | 0.011705 (rank : 77) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3SY56 | Gene names | SP6, KLF14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14). | |||||
|
SP6_MOUSE
|
||||||
| NC score | 0.011694 (rank : 78) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESX2, Q6VM22 | Gene names | Sp6, Epfn, Klf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14) (Epiprofin). | |||||
|
PITM1_HUMAN
|
||||||
| NC score | 0.011297 (rank : 79) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
RPKL1_MOUSE
|
||||||
| NC score | 0.010767 (rank : 80) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R2S1, Q8R1D4 | Gene names | Rps6kl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase-like 1 (EC 2.7.11.1). | |||||
|
SSDH_MOUSE
|
||||||
| NC score | 0.009932 (rank : 81) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWF0 | Gene names | Aldh5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
TBX21_HUMAN
|
||||||
| NC score | 0.009696 (rank : 82) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.009499 (rank : 83) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
ATS12_MOUSE
|
||||||
| NC score | 0.007759 (rank : 84) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q811B3, Q8BK92, Q8BKY1 | Gene names | Adamts12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.007504 (rank : 85) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.007350 (rank : 86) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.006663 (rank : 87) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
PRD15_HUMAN
|
||||||
| NC score | 0.005702 (rank : 88) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||
|
ZN263_HUMAN
|
||||||
| NC score | 0.003026 (rank : 89) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14978, O43387, Q96H95 | Gene names | ZNF263, FPM315 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 263 (Zinc finger protein FPM315). | |||||
|
MK15_HUMAN
|
||||||
| NC score | 0.002219 (rank : 90) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||