Please be patient as the page loads
|
SORC3_HUMAN
|
||||||
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SORC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992303 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WY21, Q59GG7, Q5JVT7, Q5JVT8, Q5VY14, Q86WQ1, Q86WQ2, Q9H1Y1, Q9H1Y2 | Gene names | SORCS1, SORCS | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (hSorCS). | |||||
|
SORC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991722 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JLC4, Q8VI45, Q922I1, Q9QY21 | Gene names | Sorcs1, Sorcs | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (mSorCS). | |||||
|
SORC2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.976781 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96PQ0, Q9P2L7 | Gene names | SORCS2, KIAA1329 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORC2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.981484 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EPR5 | Gene names | Sorcs2 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORC3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORC3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.994879 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VI51, Q9CTR4 | Gene names | Sorcs3 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORL_MOUSE
|
||||||
| θ value | 2.83479e-70 (rank : 7) | NC score | 0.460494 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
SORL_HUMAN
|
||||||
| θ value | 4.09345e-69 (rank : 8) | NC score | 0.456862 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||
|
SORT_MOUSE
|
||||||
| θ value | 6.13105e-57 (rank : 9) | NC score | 0.860466 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PHU5, Q3UHE2, Q8K043, Q9QXW6 | Gene names | Sort1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (mNTR3). | |||||
|
SORT_HUMAN
|
||||||
| θ value | 1.15626e-55 (rank : 10) | NC score | 0.859663 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99523, Q8IZ49 | Gene names | SORT1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (NT3) (Glycoprotein 95) (Gp95) (100 kDa NT receptor). | |||||
|
DEND_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.042000 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.029774 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.014169 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.017790 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
AOC3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.023359 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16853, Q45F94 | Gene names | AOC3, VAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane copper amine oxidase (EC 1.4.3.6) (Semicarbazide-sensitive amine oxidase) (SSAO) (Vascular adhesion protein 1) (VAP-1) (HPAO). | |||||
|
LTK_MOUSE
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.002433 (rank : 69) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08923 | Gene names | Ltk | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte tyrosine kinase receptor precursor (EC 2.7.10.1). | |||||
|
GPR61_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.014861 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZJ8, Q6NWS0, Q8TDV4, Q96PR4 | Gene names | GPR61, BALGR | |||
|
Domain Architecture |
|
|||||
| Description | Probable G-protein coupled receptor 61 (Biogenic amine receptor-like G-protein coupled receptor). | |||||
|
HME2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.007790 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19622 | Gene names | EN2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein engrailed-2 (Hu-En-2). | |||||
|
PAPPA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.024638 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.018746 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.051064 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
AVEN_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.030389 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQS1 | Gene names | AVEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell death regulator Aven. | |||||
|
GCH1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.048175 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30793, Q9Y4I8 | Gene names | GCH1, DYT5, GCH | |||
|
Domain Architecture |

|
|||||
| Description | GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). | |||||
|
H1X_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.030902 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92522 | Gene names | H1FX | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1x. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.016170 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
GPR61_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.012922 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 702 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C010 | Gene names | Gpr61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 61. | |||||
|
HNRPR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.008275 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43390 | Gene names | HNRPR | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein R (hnRNP R). | |||||
|
IP3KC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.014729 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96DU7, Q9UE25, Q9Y475 | Gene names | ITPKC, IP3KC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (InsP 3-kinase C) (IP3K-C). | |||||
|
ONEC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.013883 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6XBJ3 | Gene names | Onecut2, Oc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
SDK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.018902 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z5N4, Q8TEN9, Q8TEP5, Q96N44 | Gene names | SDK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.005602 (rank : 65) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.007462 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.009191 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.014634 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYN3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.013078 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZP2 | Gene names | Syn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
LSR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.014759 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86X29, O00112, O00426, Q6ZT80, Q9UQL3 | Gene names | LSR, LISCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor. | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.021699 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PRKDC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.011946 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.007694 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
S12A2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.006652 (rank : 61) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.002030 (rank : 70) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
CT012_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.017228 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NVP4, Q4F7X1, Q5QPD9, Q5QPE0, Q68DN8, Q6ZMX9, Q96NF0, Q9H1E0, Q9H442 | Gene names | C20orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf12. | |||||
|
CT012_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.012391 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C008, Q8C060 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C20orf12 homolog. | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.008801 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.006944 (rank : 60) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.004418 (rank : 66) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
GGA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.005899 (rank : 64) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GPNMB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.016715 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.006390 (rank : 63) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
PHLD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.012349 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15127 | Gene names | GPLD2, PIGPLD2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 2 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
TBCD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.006980 (rank : 59) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZP1, Q9H0B9, Q9UDD4 | Gene names | TBC1D3, PRC17 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 3 (Rab GTPase-activating protein PRC17) (Prostate cancer gene 17 protein) (TRE17 alpha protein). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.002813 (rank : 68) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MTCH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.006510 (rank : 62) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q791T5, Q3U4W7, Q791V6, Q8R0T0, Q8R1T8, Q8R2T2, Q9QZA5, Q9QZP4 | Gene names | Mtch1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier homolog 1 (Mitochondrial carrier-like protein 1). | |||||
|
PME17_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.015222 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.008305 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.004192 (rank : 67) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
VLDLR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.052722 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
CD320_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.067186 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPF0, Q53HF7 | Gene names | CD320, 8D6A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD320 antigen precursor (8D6 antigen) (FDC-signaling molecule 8D6) (FDC-SM-8D6). | |||||
|
LDLR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.051330 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35951 | Gene names | Ldlr | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
LRP12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.051376 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
LRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.051539 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
LRP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.056016 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
LRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.052346 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |
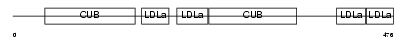
|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
LRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.050847 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75197, Q96TD6, Q9UP66 | Gene names | LRP5, LRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor. | |||||
|
LRP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.050598 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
LRP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.051154 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75581 | Gene names | LRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.051528 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88572 | Gene names | Lrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.051614 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LRP8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.051931 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
VLDLR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.054234 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98156, Q64022 | Gene names | Vldlr | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
SORC3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORC3_MOUSE
|
||||||
| NC score | 0.994879 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VI51, Q9CTR4 | Gene names | Sorcs3 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORC1_HUMAN
|
||||||
| NC score | 0.992303 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WY21, Q59GG7, Q5JVT7, Q5JVT8, Q5VY14, Q86WQ1, Q86WQ2, Q9H1Y1, Q9H1Y2 | Gene names | SORCS1, SORCS | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (hSorCS). | |||||
|
SORC1_MOUSE
|
||||||
| NC score | 0.991722 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JLC4, Q8VI45, Q922I1, Q9QY21 | Gene names | Sorcs1, Sorcs | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (mSorCS). | |||||
|
SORC2_MOUSE
|
||||||
| NC score | 0.981484 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EPR5 | Gene names | Sorcs2 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORC2_HUMAN
|
||||||
| NC score | 0.976781 (rank : 6) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96PQ0, Q9P2L7 | Gene names | SORCS2, KIAA1329 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORT_MOUSE
|
||||||
| NC score | 0.860466 (rank : 7) | θ value | 6.13105e-57 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PHU5, Q3UHE2, Q8K043, Q9QXW6 | Gene names | Sort1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (mNTR3). | |||||
|
SORT_HUMAN
|
||||||
| NC score | 0.859663 (rank : 8) | θ value | 1.15626e-55 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99523, Q8IZ49 | Gene names | SORT1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (NT3) (Glycoprotein 95) (Gp95) (100 kDa NT receptor). | |||||
|
SORL_MOUSE
|
||||||
| NC score | 0.460494 (rank : 9) | θ value | 2.83479e-70 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
SORL_HUMAN
|
||||||
| NC score | 0.456862 (rank : 10) | θ value | 4.09345e-69 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||
|
CD320_HUMAN
|
||||||
| NC score | 0.067186 (rank : 11) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPF0, Q53HF7 | Gene names | CD320, 8D6A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD320 antigen precursor (8D6 antigen) (FDC-signaling molecule 8D6) (FDC-SM-8D6). | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.056016 (rank : 12) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
VLDLR_MOUSE
|
||||||
| NC score | 0.054234 (rank : 13) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98156, Q64022 | Gene names | Vldlr | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
VLDLR_HUMAN
|
||||||
| NC score | 0.052722 (rank : 14) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
LRP3_HUMAN
|
||||||
| NC score | 0.052346 (rank : 15) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
LRP8_MOUSE
|
||||||
| NC score | 0.051931 (rank : 16) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LRP8_HUMAN
|
||||||
| NC score | 0.051614 (rank : 17) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.051539 (rank : 18) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
LRP6_MOUSE
|
||||||
| NC score | 0.051528 (rank : 19) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88572 | Gene names | Lrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP12_MOUSE
|
||||||
| NC score | 0.051376 (rank : 20) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
LDLR_MOUSE
|
||||||
| NC score | 0.051330 (rank : 21) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35951 | Gene names | Ldlr | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
LRP6_HUMAN
|
||||||
| NC score | 0.051154 (rank : 22) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75581 | Gene names | LRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.051064 (rank : 23) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
LRP5_HUMAN
|
||||||
| NC score | 0.050847 (rank : 24) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75197, Q96TD6, Q9UP66 | Gene names | LRP5, LRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor. | |||||
|
LRP5_MOUSE
|
||||||
| NC score | 0.050598 (rank : 25) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
GCH1_HUMAN
|
||||||
| NC score | 0.048175 (rank : 26) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30793, Q9Y4I8 | Gene names | GCH1, DYT5, GCH | |||
|
Domain Architecture |

|
|||||
| Description | GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). | |||||
|
DEND_HUMAN
|
||||||
| NC score | 0.042000 (rank : 27) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
H1X_HUMAN
|
||||||
| NC score | 0.030902 (rank : 28) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92522 | Gene names | H1FX | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1x. | |||||
|
AVEN_HUMAN
|
||||||
| NC score | 0.030389 (rank : 29) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQS1 | Gene names | AVEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell death regulator Aven. | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.029774 (rank : 30) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
PAPPA_HUMAN
|
||||||
| NC score | 0.024638 (rank : 31) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
AOC3_HUMAN
|
||||||
| NC score | 0.023359 (rank : 32) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16853, Q45F94 | Gene names | AOC3, VAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane copper amine oxidase (EC 1.4.3.6) (Semicarbazide-sensitive amine oxidase) (SSAO) (Vascular adhesion protein 1) (VAP-1) (HPAO). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.021699 (rank : 33) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
SDK1_HUMAN
|
||||||
| NC score | 0.018902 (rank : 34) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z5N4, Q8TEN9, Q8TEP5, Q96N44 | Gene names | SDK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.018746 (rank : 35) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.017790 (rank : 36) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
CT012_HUMAN
|
||||||
| NC score | 0.017228 (rank : 37) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NVP4, Q4F7X1, Q5QPD9, Q5QPE0, Q68DN8, Q6ZMX9, Q96NF0, Q9H1E0, Q9H442 | Gene names | C20orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf12. | |||||
|
GPNMB_HUMAN
|
||||||
| NC score | 0.016715 (rank : 38) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.016170 (rank : 39) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PME17_MOUSE
|
||||||
| NC score | 0.015222 (rank : 40) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
GPR61_HUMAN
|
||||||
| NC score | 0.014861 (rank : 41) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZJ8, Q6NWS0, Q8TDV4, Q96PR4 | Gene names | GPR61, BALGR | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 61 (Biogenic amine receptor-like G-protein coupled receptor). | |||||
|
LSR_HUMAN
|
||||||
| NC score | 0.014759 (rank : 42) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86X29, O00112, O00426, Q6ZT80, Q9UQL3 | Gene names | LSR, LISCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor. | |||||
|
IP3KC_HUMAN
|
||||||
| NC score | 0.014729 (rank : 43) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96DU7, Q9UE25, Q9Y475 | Gene names | ITPKC, IP3KC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (InsP 3-kinase C) (IP3K-C). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.014634 (rank : 44) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.014169 (rank : 45) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ONEC2_MOUSE
|
||||||
| NC score | 0.013883 (rank : 46) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6XBJ3 | Gene names | Onecut2, Oc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
SYN3_MOUSE
|
||||||
| NC score | 0.013078 (rank : 47) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZP2 | Gene names | Syn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
GPR61_MOUSE
|
||||||
| NC score | 0.012922 (rank : 48) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 702 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C010 | Gene names | Gpr61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 61. | |||||
|
CT012_MOUSE
|
||||||
| NC score | 0.012391 (rank : 49) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C008, Q8C060 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C20orf12 homolog. | |||||
|
PHLD2_HUMAN
|
||||||
| NC score | 0.012349 (rank : 50) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15127 | Gene names | GPLD2, PIGPLD2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 2 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
PRKDC_MOUSE
|
||||||
| NC score | 0.011946 (rank : 51) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.009191 (rank : 52) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.008801 (rank : 53) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.008305 (rank : 54) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
HNRPR_HUMAN
|
||||||
| NC score | 0.008275 (rank : 55) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43390 | Gene names | HNRPR | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein R (hnRNP R). | |||||
|
HME2_HUMAN
|
||||||
| NC score | 0.007790 (rank : 56) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19622 | Gene names | EN2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein engrailed-2 (Hu-En-2). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.007694 (rank : 57) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.007462 (rank : 58) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
TBCD3_HUMAN
|
||||||
| NC score | 0.006980 (rank : 59) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZP1, Q9H0B9, Q9UDD4 | Gene names | TBC1D3, PRC17 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 3 (Rab GTPase-activating protein PRC17) (Prostate cancer gene 17 protein) (TRE17 alpha protein). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.006944 (rank : 60) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
S12A2_MOUSE
|
||||||
| NC score | 0.006652 (rank : 61) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
MTCH1_MOUSE
|
||||||
| NC score | 0.006510 (rank : 62) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q791T5, Q3U4W7, Q791V6, Q8R0T0, Q8R1T8, Q8R2T2, Q9QZA5, Q9QZP4 | Gene names | Mtch1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier homolog 1 (Mitochondrial carrier-like protein 1). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.006390 (rank : 63) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.005899 (rank : 64) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.005602 (rank : 65) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.004418 (rank : 66) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.004192 (rank : 67) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.002813 (rank : 68) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LTK_MOUSE
|
||||||
| NC score | 0.002433 (rank : 69) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08923 | Gene names | Ltk | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte tyrosine kinase receptor precursor (EC 2.7.10.1). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.002030 (rank : 70) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||