Please be patient as the page loads
|
MMP15_MOUSE
|
||||||
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MMP15_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996872 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51511, Q14111 | Gene names | MMP15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP) (SMCP- 2). | |||||
|
MMP15_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
MMP16_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.994020 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51512, Q14824, Q52H48 | Gene names | MMP16, MMPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP) (MMP- X2). | |||||
|
MMP16_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.993920 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
MMP24_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.994072 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y5R2, Q9H440 | Gene names | MMP24, MT5MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP). | |||||
|
MMP14_HUMAN
|
||||||
| θ value | 3.1398e-178 (rank : 6) | NC score | 0.994454 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50281, Q92678 | Gene names | MMP14 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP- X1). | |||||
|
MMP14_MOUSE
|
||||||
| θ value | 1.19313e-177 (rank : 7) | NC score | 0.994434 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P53690, O08645, O35369 | Gene names | Mmp14, Mtmmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP-X1) (MT-MMP). | |||||
|
MMP24_MOUSE
|
||||||
| θ value | 2.32852e-173 (rank : 8) | NC score | 0.991665 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R0S2, Q9Z0J9 | Gene names | Mmp24, Mmp21, Mt5mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) (MMP-21). | |||||
|
MMP25_HUMAN
|
||||||
| θ value | 5.67233e-95 (rank : 9) | NC score | 0.983777 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
MMP17_MOUSE
|
||||||
| θ value | 3.67668e-94 (rank : 10) | NC score | 0.981034 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP17_HUMAN
|
||||||
| θ value | 3.22093e-90 (rank : 11) | NC score | 0.980362 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP8_HUMAN
|
||||||
| θ value | 2.0926e-81 (rank : 12) | NC score | 0.968605 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P22894, Q45F99 | Gene names | MMP8, CLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (PMNL collagenase) (PMNL-CL). | |||||
|
MMP8_MOUSE
|
||||||
| θ value | 1.95861e-79 (rank : 13) | NC score | 0.969381 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O70138, O88733 | Gene names | Mmp8 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (Collagenase 2). | |||||
|
MMP12_HUMAN
|
||||||
| θ value | 2.1655e-78 (rank : 14) | NC score | 0.969472 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P39900 | Gene names | MMP12, HME | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (HME) (Matrix metalloproteinase-12) (MMP-12) (Macrophage elastase) (ME). | |||||
|
MMP1_HUMAN
|
||||||
| θ value | 4.08394e-77 (rank : 15) | NC score | 0.964025 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
MMP11_MOUSE
|
||||||
| θ value | 1.5519e-76 (rank : 16) | NC score | 0.976201 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
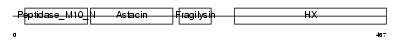
MMP10_MOUSE
|
||||||
| θ value | 1.00591e-75 (rank : 17) | NC score | 0.964787 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |
|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP3_MOUSE
|
||||||
| θ value | 1.31376e-75 (rank : 18) | NC score | 0.964061 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
MMP13_HUMAN
|
||||||
| θ value | 2.92676e-75 (rank : 19) | NC score | 0.966002 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP13_MOUSE
|
||||||
| θ value | 1.22964e-73 (rank : 20) | NC score | 0.964638 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP3_HUMAN
|
||||||
| θ value | 2.09745e-73 (rank : 21) | NC score | 0.963260 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP11_HUMAN
|
||||||
| θ value | 2.31901e-72 (rank : 22) | NC score | 0.976448 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP10_HUMAN
|
||||||
| θ value | 5.71191e-71 (rank : 23) | NC score | 0.962679 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP1A_MOUSE
|
||||||
| θ value | 1.66191e-70 (rank : 24) | NC score | 0.964364 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9EPL5 | Gene names | Mmp1a, McolA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase A precursor (EC 3.4.24.7) (Matrix metalloproteinase-1a) (MMP-1a) (Mcol-A). | |||||
|
MMP20_HUMAN
|
||||||
| θ value | 2.24614e-67 (rank : 25) | NC score | 0.965312 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60882, Q6DKT9 | Gene names | MMP20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP20_MOUSE
|
||||||
| θ value | 2.4834e-66 (rank : 26) | NC score | 0.964976 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P57748 | Gene names | Mmp20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP12_MOUSE
|
||||||
| θ value | 4.68348e-65 (rank : 27) | NC score | 0.964840 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P34960, Q8BJ92, Q8BJB3, Q8BJB6, Q8BJB8, Q8BJB9, Q8VED6 | Gene names | Mmp12, Mme, Mmel | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (MME) (Matrix metalloproteinase-12) (MMP-12). | |||||
|
MMP1B_MOUSE
|
||||||
| θ value | 6.11685e-65 (rank : 28) | NC score | 0.962037 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9EPL6 | Gene names | Mmp1b, McolB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase B precursor (EC 3.4.24.7) (Matrix metalloproteinase-1b) (MMP-1b) (Mcol-B). | |||||
|
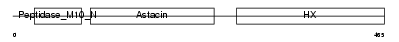
MMP19_HUMAN
|
||||||
| θ value | 1.84173e-61 (rank : 29) | NC score | 0.964934 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99542, O15278, O95606, Q99580 | Gene names | MMP19, MMP18, RASI | |||
|
Domain Architecture |
|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI) (MMP-18). | |||||
|
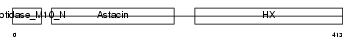
MMP19_MOUSE
|
||||||
| θ value | 5.35859e-61 (rank : 30) | NC score | 0.966815 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JHI0, Q8C360 | Gene names | Mmp19, Rasi | |||
|
Domain Architecture |
|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI). | |||||
|
MMP28_HUMAN
|
||||||
| θ value | 8.55514e-59 (rank : 31) | NC score | 0.973971 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H239, Q96TE2 | Gene names | MMP28, MMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-28 precursor (EC 3.4.24.-) (MMP-28) (Epilysin). | |||||
|
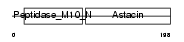
MMP7_HUMAN
|
||||||
| θ value | 5.7518e-47 (rank : 32) | NC score | 0.954144 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP7_MOUSE
|
||||||
| θ value | 7.03111e-45 (rank : 33) | NC score | 0.951716 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |
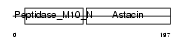
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP26_HUMAN
|
||||||
| θ value | 2.67802e-36 (rank : 34) | NC score | 0.949525 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |
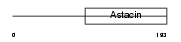
|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
MMP2_HUMAN
|
||||||
| θ value | 2.77131e-33 (rank : 35) | NC score | 0.879240 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
MMP2_MOUSE
|
||||||
| θ value | 1.0531e-32 (rank : 36) | NC score | 0.879380 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
MMP21_HUMAN
|
||||||
| θ value | 8.91499e-32 (rank : 37) | NC score | 0.919727 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8N119, Q8NG02 | Gene names | MMP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MMP21_MOUSE
|
||||||
| θ value | 4.89182e-30 (rank : 38) | NC score | 0.918948 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K3F2 | Gene names | Mmp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MMP9_MOUSE
|
||||||
| θ value | 5.97985e-28 (rank : 39) | NC score | 0.829130 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
MMP9_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 40) | NC score | 0.839129 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
VTNC_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 41) | NC score | 0.741363 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
VTNC_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 42) | NC score | 0.712949 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 43) | NC score | 0.384901 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
HEMO_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 44) | NC score | 0.661263 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P02790 | Gene names | HPX | |||
|
Domain Architecture |
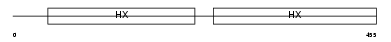
|
|||||
| Description | Hemopexin precursor (Beta-1B-glycoprotein). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 45) | NC score | 0.316123 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
HEMO_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 46) | NC score | 0.584965 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91X72, P97824, Q8WUP0 | Gene names | Hpx, Hpxn | |||
|
Domain Architecture |
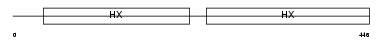
|
|||||
| Description | Hemopexin precursor. | |||||
|
BCL7C_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 47) | NC score | 0.037675 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 48) | NC score | 0.060797 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 49) | NC score | 0.019354 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.012466 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.029380 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.21417 (rank : 52) | NC score | 0.011615 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 53) | NC score | -0.000269 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.028425 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
BAT3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.030317 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.032185 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.006976 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
RS3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.026262 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62908, P17073, P47933 | Gene names | Rps3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 40S ribosomal protein S3. | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.030173 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.014545 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
RS3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.025564 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23396, Q498B5, Q8NI95 | Gene names | RPS3 | |||
|
Domain Architecture |
|
|||||
| Description | 40S ribosomal protein S3. | |||||
|
MELPH_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.019681 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
ATS20_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.010131 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59510 | Gene names | ADAMTS20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
COG8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.013929 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.012038 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.017128 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.011130 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
INP5E_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.014964 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.016625 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
ATS13_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.043980 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.022333 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.013194 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.013235 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.013252 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.006606 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.002954 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | -0.005104 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.006598 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | -0.001906 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.003721 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.027322 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
BIM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.014057 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43521, O43522 | Gene names | BCL2L11, BIM | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.012923 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.007267 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
BIM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.013025 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54918, O54919, O54920 | Gene names | Bcl2l11, Bim | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
CBPZ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.001981 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
FOXJ2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.004403 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.009243 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MSH5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.004834 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.013518 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SDC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.007923 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18828, Q62278, Q9WVD2 | Gene names | Sdc1, Synd-1, Synd1 | |||
|
Domain Architecture |
|
|||||
| Description | Syndecan-1 precursor (SYND1). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.006652 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.009473 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.003948 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
ABR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.001078 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
ATS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.015515 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
ATS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.014998 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C9W3 | Gene names | Adamts2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.012418 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
IDD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.002776 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.008184 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.002708 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.006144 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
SEL1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.062338 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.060941 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
MMP15_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
MMP15_HUMAN
|
||||||
| NC score | 0.996872 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51511, Q14111 | Gene names | MMP15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP) (SMCP- 2). | |||||
|
MMP14_HUMAN
|
||||||
| NC score | 0.994454 (rank : 3) | θ value | 3.1398e-178 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50281, Q92678 | Gene names | MMP14 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP- X1). | |||||
|
MMP14_MOUSE
|
||||||
| NC score | 0.994434 (rank : 4) | θ value | 1.19313e-177 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P53690, O08645, O35369 | Gene names | Mmp14, Mtmmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP-X1) (MT-MMP). | |||||
|
MMP24_HUMAN
|
||||||
| NC score | 0.994072 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y5R2, Q9H440 | Gene names | MMP24, MT5MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP). | |||||
|
MMP16_HUMAN
|
||||||
| NC score | 0.994020 (rank : 6) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51512, Q14824, Q52H48 | Gene names | MMP16, MMPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP) (MMP- X2). | |||||
|
MMP16_MOUSE
|
||||||
| NC score | 0.993920 (rank : 7) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
MMP24_MOUSE
|
||||||
| NC score | 0.991665 (rank : 8) | θ value | 2.32852e-173 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R0S2, Q9Z0J9 | Gene names | Mmp24, Mmp21, Mt5mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) (MMP-21). | |||||
|
MMP25_HUMAN
|
||||||
| NC score | 0.983777 (rank : 9) | θ value | 5.67233e-95 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
MMP17_MOUSE
|
||||||
| NC score | 0.981034 (rank : 10) | θ value | 3.67668e-94 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP17_HUMAN
|
||||||
| NC score | 0.980362 (rank : 11) | θ value | 3.22093e-90 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP11_HUMAN
|
||||||
| NC score | 0.976448 (rank : 12) | θ value | 2.31901e-72 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP11_MOUSE
|
||||||
| NC score | 0.976201 (rank : 13) | θ value | 1.5519e-76 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP28_HUMAN
|
||||||
| NC score | 0.973971 (rank : 14) | θ value | 8.55514e-59 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H239, Q96TE2 | Gene names | MMP28, MMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-28 precursor (EC 3.4.24.-) (MMP-28) (Epilysin). | |||||
|
MMP12_HUMAN
|
||||||
| NC score | 0.969472 (rank : 15) | θ value | 2.1655e-78 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P39900 | Gene names | MMP12, HME | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (HME) (Matrix metalloproteinase-12) (MMP-12) (Macrophage elastase) (ME). | |||||
|
MMP8_MOUSE
|
||||||
| NC score | 0.969381 (rank : 16) | θ value | 1.95861e-79 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O70138, O88733 | Gene names | Mmp8 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (Collagenase 2). | |||||
|
MMP8_HUMAN
|
||||||
| NC score | 0.968605 (rank : 17) | θ value | 2.0926e-81 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P22894, Q45F99 | Gene names | MMP8, CLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (PMNL collagenase) (PMNL-CL). | |||||
|
MMP19_MOUSE
|
||||||
| NC score | 0.966815 (rank : 18) | θ value | 5.35859e-61 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JHI0, Q8C360 | Gene names | Mmp19, Rasi | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI). | |||||
|
MMP13_HUMAN
|
||||||
| NC score | 0.966002 (rank : 19) | θ value | 2.92676e-75 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP20_HUMAN
|
||||||
| NC score | 0.965312 (rank : 20) | θ value | 2.24614e-67 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60882, Q6DKT9 | Gene names | MMP20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP20_MOUSE
|
||||||
| NC score | 0.964976 (rank : 21) | θ value | 2.4834e-66 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P57748 | Gene names | Mmp20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP19_HUMAN
|
||||||
| NC score | 0.964934 (rank : 22) | θ value | 1.84173e-61 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99542, O15278, O95606, Q99580 | Gene names | MMP19, MMP18, RASI | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI) (MMP-18). | |||||
|
MMP12_MOUSE
|
||||||
| NC score | 0.964840 (rank : 23) | θ value | 4.68348e-65 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P34960, Q8BJ92, Q8BJB3, Q8BJB6, Q8BJB8, Q8BJB9, Q8VED6 | Gene names | Mmp12, Mme, Mmel | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (MME) (Matrix metalloproteinase-12) (MMP-12). | |||||
|
MMP10_MOUSE
|
||||||
| NC score | 0.964787 (rank : 24) | θ value | 1.00591e-75 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP13_MOUSE
|
||||||
| NC score | 0.964638 (rank : 25) | θ value | 1.22964e-73 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP1A_MOUSE
|
||||||
| NC score | 0.964364 (rank : 26) | θ value | 1.66191e-70 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9EPL5 | Gene names | Mmp1a, McolA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase A precursor (EC 3.4.24.7) (Matrix metalloproteinase-1a) (MMP-1a) (Mcol-A). | |||||
|
MMP3_MOUSE
|
||||||
| NC score | 0.964061 (rank : 27) | θ value | 1.31376e-75 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
MMP1_HUMAN
|
||||||
| NC score | 0.964025 (rank : 28) | θ value | 4.08394e-77 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
MMP3_HUMAN
|
||||||
| NC score | 0.963260 (rank : 29) | θ value | 2.09745e-73 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP10_HUMAN
|
||||||
| NC score | 0.962679 (rank : 30) | θ value | 5.71191e-71 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP1B_MOUSE
|
||||||
| NC score | 0.962037 (rank : 31) | θ value | 6.11685e-65 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9EPL6 | Gene names | Mmp1b, McolB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase B precursor (EC 3.4.24.7) (Matrix metalloproteinase-1b) (MMP-1b) (Mcol-B). | |||||
|
MMP7_HUMAN
|
||||||
| NC score | 0.954144 (rank : 32) | θ value | 5.7518e-47 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP7_MOUSE
|
||||||
| NC score | 0.951716 (rank : 33) | θ value | 7.03111e-45 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP26_HUMAN
|
||||||
| NC score | 0.949525 (rank : 34) | θ value | 2.67802e-36 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
MMP21_HUMAN
|
||||||
| NC score | 0.919727 (rank : 35) | θ value | 8.91499e-32 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8N119, Q8NG02 | Gene names | MMP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MMP21_MOUSE
|
||||||
| NC score | 0.918948 (rank : 36) | θ value | 4.89182e-30 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K3F2 | Gene names | Mmp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MMP2_MOUSE
|
||||||
| NC score | 0.879380 (rank : 37) | θ value | 1.0531e-32 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
MMP2_HUMAN
|
||||||
| NC score | 0.879240 (rank : 38) | θ value | 2.77131e-33 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
MMP9_HUMAN
|
||||||
| NC score | 0.839129 (rank : 39) | θ value | 3.87602e-27 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
MMP9_MOUSE
|
||||||
| NC score | 0.829130 (rank : 40) | θ value | 5.97985e-28 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
VTNC_MOUSE
|
||||||
| NC score | 0.741363 (rank : 41) | θ value | 4.44505e-15 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
VTNC_HUMAN
|
||||||
| NC score | 0.712949 (rank : 42) | θ value | 1.58096e-12 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
HEMO_HUMAN
|
||||||
| NC score | 0.661263 (rank : 43) | θ value | 7.59969e-07 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P02790 | Gene names | HPX | |||
|
Domain Architecture |

|
|||||
| Description | Hemopexin precursor (Beta-1B-glycoprotein). | |||||
|
HEMO_MOUSE
|
||||||
| NC score | 0.584965 (rank : 44) | θ value | 0.000602161 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91X72, P97824, Q8WUP0 | Gene names | Hpx, Hpxn | |||
|
Domain Architecture |

|
|||||
| Description | Hemopexin precursor. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.384901 (rank : 45) | θ value | 1.38499e-08 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.316123 (rank : 46) | θ value | 5.44631e-05 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SEL1L_HUMAN
|
||||||
| NC score | 0.062338 (rank : 47) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| NC score | 0.060941 (rank : 48) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.060797 (rank : 49) | θ value | 0.0563607 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
ATS13_MOUSE
|
||||||
| NC score | 0.043980 (rank : 50) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
BCL7C_MOUSE
|
||||||
| NC score | 0.037675 (rank : 51) | θ value | 0.0252991 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.032185 (rank : 52) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
BAT3_HUMAN
|
||||||
| NC score | 0.030317 (rank : 53) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.030173 (rank : 54) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.029380 (rank : 55) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.028425 (rank : 56) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.027322 (rank : 57) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
RS3_MOUSE
|
||||||
| NC score | 0.026262 (rank : 58) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62908, P17073, P47933 | Gene names | Rps3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 40S ribosomal protein S3. | |||||
|
RS3_HUMAN
|
||||||
| NC score | 0.025564 (rank : 59) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23396, Q498B5, Q8NI95 | Gene names | RPS3 | |||
|
Domain Architecture |

|
|||||
| Description | 40S ribosomal protein S3. | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.022333 (rank : 60) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
MELPH_MOUSE
|
||||||
| NC score | 0.019681 (rank : 61) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.019354 (rank : 62) | θ value | 0.0961366 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.017128 (rank : 63) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.016625 (rank : 64) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
ATS2_HUMAN
|
||||||
| NC score | 0.015515 (rank : 65) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
ATS2_MOUSE
|
||||||
| NC score | 0.014998 (rank : 66) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C9W3 | Gene names | Adamts2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
INP5E_HUMAN
|
||||||
| NC score | 0.014964 (rank : 67) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.014545 (rank : 68) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
BIM_HUMAN
|
||||||
| NC score | 0.014057 (rank : 69) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43521, O43522 | Gene names | BCL2L11, BIM | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
COG8_MOUSE
|
||||||
| NC score | 0.013929 (rank : 70) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.013518 (rank : 71) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.013252 (rank : 72) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.013235 (rank : 73) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.013194 (rank : 74) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
BIM_MOUSE
|
||||||
| NC score | 0.013025 (rank : 75) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54918, O54919, O54920 | Gene names | Bcl2l11, Bim | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.012923 (rank : 76) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.012466 (rank : 77) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.012418 (rank : 78) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.012038 (rank : 79) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.011615 (rank : 80) | θ value | 0.21417 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.011130 (rank : 81) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
ATS20_HUMAN
|
||||||
| NC score | 0.010131 (rank : 82) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59510 | Gene names | ADAMTS20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.009473 (rank : 83) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.009243 (rank : 84) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.008184 (rank : 85) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
SDC1_MOUSE
|
||||||
| NC score | 0.007923 (rank : 86) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18828, Q62278, Q9WVD2 | Gene names | Sdc1, Synd-1, Synd1 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-1 precursor (SYND1). | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.007267 (rank : 87) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.006976 (rank : 88) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.006652 (rank : 89) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.006606 (rank : 90) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.006598 (rank : 91) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.006144 (rank : 92) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
MSH5_HUMAN
|
||||||
| NC score | 0.004834 (rank : 93) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||
|
FOXJ2_HUMAN
|
||||||
| NC score | 0.004403 (rank : 94) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.003948 (rank : 95) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.003721 (rank : 96) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.002954 (rank : 97) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
IDD_MOUSE
|
||||||
| NC score | 0.002776 (rank : 98) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.002708 (rank : 99) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CBPZ_MOUSE
|
||||||
| NC score | 0.001981 (rank : 100) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.001078 (rank : 101) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | -0.000269 (rank : 102) | θ value | 0.279714 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | -0.001906 (rank : 103) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | -0.005104 (rank : 104) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||