Please be patient as the page loads
|
IC1_MOUSE
|
||||||
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IC1_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 164 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
IC1_HUMAN
|
||||||
| θ value | 2.10118e-182 (rank : 2) | NC score | 0.990168 (rank : 2) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P05155, Q547W3, Q59EI5, Q96FE0 | Gene names | SERPING1, C1IN, C1NH | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
A2AP_MOUSE
|
||||||
| θ value | 1.3722e-40 (rank : 3) | NC score | 0.954778 (rank : 3) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61247 | Gene names | Serpinf2, Pli | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-antiplasmin precursor (Alpha-2-plasmin inhibitor) (Alpha-2-PI) (Alpha-2-AP). | |||||
|
A2AP_HUMAN
|
||||||
| θ value | 2.86122e-38 (rank : 4) | NC score | 0.954354 (rank : 4) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P08697 | Gene names | SERPINF2, AAP, PLI | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-antiplasmin precursor (Alpha-2-plasmin inhibitor) (Alpha-2-PI) (Alpha-2-AP). | |||||
|
SPA3K_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 5) | NC score | 0.936726 (rank : 12) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P07759, Q91W80, Q91X80 | Gene names | Serpina3k, Mcm2, Spi2 | |||
|
Domain Architecture |
|
|||||
| Description | Serine protease inhibitor A3K precursor (Contrapsin) (Serpin A3K). | |||||
|
SPA3M_MOUSE
|
||||||
| θ value | 1.79631e-32 (rank : 6) | NC score | 0.936112 (rank : 15) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q03734, Q62260 | Gene names | Serpina3m | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3M precursor (Serpin A3M). | |||||
|
AACT_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 7) | NC score | 0.939834 (rank : 8) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P01011, Q13703, Q59GP9, Q6LBY8, Q6LDT7, Q6NSC9, Q8N177, Q96DW8, Q9UNU9 | Gene names | SERPINA3, AACT | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1-antichymotrypsin precursor (ACT) [Contains: Alpha-1- antichymotrypsin His-Pro-less]. | |||||
|
PEDF_HUMAN
|
||||||
| θ value | 1.6813e-30 (rank : 8) | NC score | 0.948485 (rank : 6) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P36955, Q13236, Q96CT1, Q96R01, Q9BWA4 | Gene names | SERPINF1, PEDF | |||
|
Domain Architecture |
|
|||||
| Description | Pigment epithelium-derived factor precursor (PEDF) (Serpin-F1) (EPC- 1). | |||||
|
PEDF_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 9) | NC score | 0.949033 (rank : 5) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P97298, O70629, O88691 | Gene names | Serpinf1, Pedf, Sdf3 | |||
|
Domain Architecture |
|
|||||
| Description | Pigment epithelium-derived factor precursor (PEDF) (Serpin-F1) (Stromal cell-derived factor 3) (SDF-3) (Caspin). | |||||
|
SPA3C_MOUSE
|
||||||
| θ value | 4.14116e-29 (rank : 10) | NC score | 0.936566 (rank : 13) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P29621 | Gene names | Serpina3c, Klkbp | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor A3C precursor (Kallikrein-binding protein) (KBP) (Serpin A3C). | |||||
|
SPA3N_MOUSE
|
||||||
| θ value | 1.73987e-27 (rank : 11) | NC score | 0.932994 (rank : 24) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q91WP6, Q62258 | Gene names | Serpina3n, Spi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3N precursor (Serpin A3N). | |||||
|
SPA3F_MOUSE
|
||||||
| θ value | 6.61148e-27 (rank : 12) | NC score | 0.933984 (rank : 20) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q80X76 | Gene names | Serpina3f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3F (Serpin A3F). | |||||
|
IPSP_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 13) | NC score | 0.936467 (rank : 14) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P05154, Q07616, Q9UG30 | Gene names | SERPINA5, PCI, PLANH3, PROCI | |||
|
Domain Architecture |
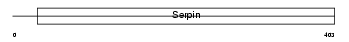
|
|||||
| Description | Plasma serine protease inhibitor precursor (PCI) (Protein C inhibitor) (Serpin A5) (Plasminogen activator inhibitor 3) (PAI-3) (PAI3) (Acrosomal serine protease inhibitor). | |||||
|
SPA3B_MOUSE
|
||||||
| θ value | 1.92365e-26 (rank : 14) | NC score | 0.933728 (rank : 21) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8BYY9 | Gene names | Serpina3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3B precursor (Serpin A3B). | |||||
|
SPA3G_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 15) | NC score | 0.934305 (rank : 19) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q5I2A0, Q6PG99 | Gene names | Serpina3g, Spi2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3G (Serine protease inhibitor 2A) (Serpin A3G) (Serpin 2A). | |||||
|
SPB3_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 16) | NC score | 0.926015 (rank : 45) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P29508, Q96J21 | Gene names | SERPINB3, SCCA, SCCA1 | |||
|
Domain Architecture |
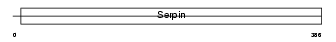
|
|||||
| Description | Serpin B3 (Squamous cell carcinoma antigen 1) (SCCA-1) (Protein T4-A). | |||||
|
SPI2_MOUSE
|
||||||
| θ value | 2.12685e-25 (rank : 17) | NC score | 0.938159 (rank : 9) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9JK88, Q9D8Z3, Q9D955 | Gene names | Serpini2, Spi14 | |||
|
Domain Architecture |
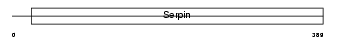
|
|||||
| Description | Serpin I2 precursor (Serine protease inhibitor 14). | |||||
|
SPI2_HUMAN
|
||||||
| θ value | 3.62785e-25 (rank : 18) | NC score | 0.937915 (rank : 10) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O75830 | Gene names | SERPINI2, MEPI, PI14 | |||
|
Domain Architecture |
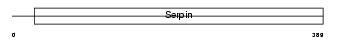
|
|||||
| Description | Serpin I2 precursor (Myoepithelium-derived serine protease inhibitor) (Pancpin) (Protease inhibitor 14) (TSA2004). | |||||
|
A1AT5_MOUSE
|
||||||
| θ value | 6.18819e-25 (rank : 19) | NC score | 0.929126 (rank : 32) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q00898 | Gene names | Serpina1e, Dom5, Spi1-5 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1-antitrypsin 1-5 precursor (Serine protease inhibitor 1-5) (Alpha-1 protease inhibitor 5). | |||||
|
SPB8_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 20) | NC score | 0.920331 (rank : 58) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P50452 | Gene names | SERPINB8, PI8 | |||
|
Domain Architecture |
|
|||||
| Description | Serpin B8 (Cytoplasmic antiproteinase 2) (CAP-2) (CAP2) (Protease inhibitor 8). | |||||
|
SPB4_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 21) | NC score | 0.924028 (rank : 50) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P48594 | Gene names | SERPINB4, SCCA2 | |||
|
Domain Architecture |
|
|||||
| Description | Serpin B4 (Squamous cell carcinoma antigen 2) (SCCA-2) (Leupin). | |||||
|
SPB8_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 22) | NC score | 0.921646 (rank : 53) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O08800 | Gene names | Serpinb8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B8. | |||||
|
KAIN_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 23) | NC score | 0.928508 (rank : 34) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P29622, Q53XB5, Q86TR9, Q96BZ5 | Gene names | SERPINA4, KST, PI4 | |||
|
Domain Architecture |
|
|||||
| Description | Kallistatin precursor (Serpin A4) (Kallikrein inhibitor) (Protease inhibitor 4). | |||||
|
IPSP_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 24) | NC score | 0.933317 (rank : 23) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P70458 | Gene names | Serpina5, Pci | |||
|
Domain Architecture |
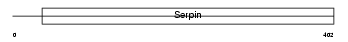
|
|||||
| Description | Plasma serine protease inhibitor precursor (PCI) (Protein C inhibitor) (Serpin A5) (Plasminogen activator inhibitor 3) (PAI-3) (PAI3). | |||||
|
SPA9_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 25) | NC score | 0.931487 (rank : 28) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9D7D2 | Gene names | Serpina9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A9 precursor. | |||||
|
ANT3_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 26) | NC score | 0.931894 (rank : 26) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P01008, P78439, P78447, Q13815, Q7KZ43 | Gene names | SERPINC1, AT3 | |||
|
Domain Architecture |
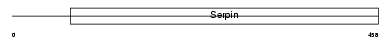
|
|||||
| Description | Antithrombin-III precursor (ATIII). | |||||
|
NEUS_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 27) | NC score | 0.934766 (rank : 18) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q99574, Q6AHZ4 | Gene names | SERPINI1, PI12 | |||
|
Domain Architecture |
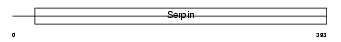
|
|||||
| Description | Neuroserpin precursor (Serpin I1) (Protease inhibitor 12). | |||||
|
A1AT2_MOUSE
|
||||||
| θ value | 1.5242e-23 (rank : 28) | NC score | 0.925189 (rank : 46) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P22599, Q61283, Q80ZH5, Q8VC20 | Gene names | Serpina1b, Aat2, Dom2, Spi1-2 | |||
|
Domain Architecture |
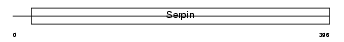
|
|||||
| Description | Alpha-1-antitrypsin 1-2 precursor (Serine protease inhibitor 1-2) (Alpha-1 protease inhibitor 2) (Alpha-1-antiproteinase) (AAT). | |||||
|
PAI2_MOUSE
|
||||||
| θ value | 1.5242e-23 (rank : 29) | NC score | 0.924875 (rank : 48) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P12388, O35687, Q9QWP6, Q9QWP7, Q9QWP8, Q9QWP9, Q9QWQ0, Q9QWZ5, Q9QWZ6 | Gene names | Serpinb2, Pai2, Planh2 | |||
|
Domain Architecture |
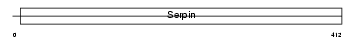
|
|||||
| Description | Plasminogen activator inhibitor 2, macrophage (PAI-2). | |||||
|
A1AT3_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 30) | NC score | 0.925038 (rank : 47) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q00896 | Gene names | Serpina1c, Dom3, Spi1-3 | |||
|
Domain Architecture |
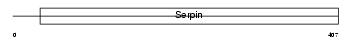
|
|||||
| Description | Alpha-1-antitrypsin 1-3 precursor (Serine protease inhibitor 1-3) (Alpha-1 protease inhibitor 3). | |||||
|
A1AT4_MOUSE
|
||||||
| θ value | 3.39556e-23 (rank : 31) | NC score | 0.928470 (rank : 35) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q00897 | Gene names | Serpina1d, Dom4, Spi1-4 | |||
|
Domain Architecture |
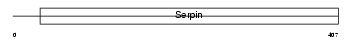
|
|||||
| Description | Alpha-1-antitrypsin 1-4 precursor (Serine protease inhibitor 1-4) (Alpha-1 protease inhibitor 4). | |||||
|
A1AT_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 32) | NC score | 0.931315 (rank : 29) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P01009, Q13672, Q5U0M1, Q96BF9, Q96ES1, Q9P1P0 | Gene names | SERPINA1, AAT, PI | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin precursor (Alpha-1 protease inhibitor) (Alpha-1- antiproteinase). | |||||
|
ANT3_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 33) | NC score | 0.932544 (rank : 25) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P32261 | Gene names | Serpinc1, At3 | |||
|
Domain Architecture |
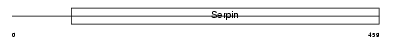
|
|||||
| Description | Antithrombin-III precursor (ATIII). | |||||
|
A1AT1_MOUSE
|
||||||
| θ value | 5.79196e-23 (rank : 34) | NC score | 0.928049 (rank : 37) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P07758, Q80YB8, Q8JZV6, Q91XB8 | Gene names | Serpina1a, Dom1, Spi1-1 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1-antitrypsin 1-1 precursor (Serine protease inhibitor 1-1) (Alpha-1 protease inhibitor 1) (Alpha-1-antiproteinase) (AAT). | |||||
|
A1AT6_MOUSE
|
||||||
| θ value | 9.87957e-23 (rank : 35) | NC score | 0.924382 (rank : 49) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P81105, Q91V74, Q91WH5, Q91XC1 | Gene names | Spi1-6, Dom6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1-antitrypsin 1-6 precursor (Serine protease inhibitor 1-6) (Alpha-1 protease inhibitor 6). | |||||
|
THBG_HUMAN
|
||||||
| θ value | 9.87957e-23 (rank : 36) | NC score | 0.930208 (rank : 30) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P05543 | Gene names | SERPINA7, TBG | |||
|
Domain Architecture |
|
|||||
| Description | Thyroxine-binding globulin precursor (T4-binding globulin) (Serpin A7). | |||||
|
PAI2_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 37) | NC score | 0.919110 (rank : 62) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P05120, Q96E96 | Gene names | SERPINB2, PAI2, PLANH2 | |||
|
Domain Architecture |
|
|||||
| Description | Plasminogen activator inhibitor 2 precursor (PAI-2) (Placental plasminogen activator inhibitor) (Monocyte Arg-serpin) (Urokinase inhibitor). | |||||
|
SPA3A_MOUSE
|
||||||
| θ value | 6.40375e-22 (rank : 38) | NC score | 0.928727 (rank : 33) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q6P4P1, Q9D490 | Gene names | Serpina3a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3A precursor (Serpin A3A). | |||||
|
ZPI_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 39) | NC score | 0.939904 (rank : 7) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UK55, Q6UWX9, Q86U20 | Gene names | SERPINA10, ZPI | |||
|
Domain Architecture |

|
|||||
| Description | Protein Z-dependent protease inhibitor precursor (PZ-dependent protease inhibitor) (PZI) (Serpin A10). | |||||
|
SPA11_MOUSE
|
||||||
| θ value | 1.86321e-21 (rank : 40) | NC score | 0.926175 (rank : 43) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8CIE0 | Gene names | Serpina11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A11 precursor. | |||||
|
SPA11_HUMAN
|
||||||
| θ value | 2.43343e-21 (rank : 41) | NC score | 0.926552 (rank : 42) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q86U17 | Gene names | SERPINA11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A11 precursor. | |||||
|
SPA12_MOUSE
|
||||||
| θ value | 3.17815e-21 (rank : 42) | NC score | 0.935776 (rank : 16) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q7TMF5, Q9CQ32 | Gene names | Serpina12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A12 precursor (Visceral adipose-specific serpin) (Visceral adipose tissue-derived serine protease inhibitor) (Vaspin). | |||||
|
SPB6_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 43) | NC score | 0.917450 (rank : 65) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P35237, Q96J44 | Gene names | SERPINB6, PI6, PTI | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B6 (Placental thrombin inhibitor) (Cytoplasmic antiproteinase) (CAP) (Protease inhibitor 6) (PI-6). | |||||
|
ZPI_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 44) | NC score | 0.937638 (rank : 11) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8R121, Q8VCV8 | Gene names | Serpina10, Zpi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Z-dependent protease inhibitor precursor (PZ-dependent protease inhibitor) (PZI) (Serpin A10). | |||||
|
NEUS_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 45) | NC score | 0.927556 (rank : 39) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O35684, Q543F7, Q922U6 | Gene names | Serpini1, Pi12, Spi17 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroserpin precursor (Serpin I1) (Protease inhibitor 17). | |||||
|
SPB9_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 46) | NC score | 0.923306 (rank : 51) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P50453 | Gene names | SERPINB9, PI9 | |||
|
Domain Architecture |
|
|||||
| Description | Serpin B9 (Cytoplasmic antiproteinase 3) (CAP-3) (CAP3) (Protease inhibitor 9). | |||||
|
CBG_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 47) | NC score | 0.933318 (rank : 22) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P08185, Q7Z2Q9 | Gene names | SERPINA6, CBG | |||
|
Domain Architecture |
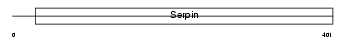
|
|||||
| Description | Corticosteroid-binding globulin precursor (CBG) (Transcortin) (Serpin A6). | |||||
|
SPB12_MOUSE
|
||||||
| θ value | 3.51386e-20 (rank : 48) | NC score | 0.917734 (rank : 64) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9D7P9, Q6UKZ3 | Gene names | Serpinb12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B12. | |||||
|
ILEU_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 49) | NC score | 0.921552 (rank : 55) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P30740, Q53FB9, Q5W0E1 | Gene names | SERPINB1, ELANH2, PI2 | |||
|
Domain Architecture |
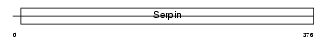
|
|||||
| Description | Leukocyte elastase inhibitor (LEI) (Serpin B1) (Monocyte/neutrophil elastase inhibitor) (M/NEI) (EI). | |||||
|
PAI1_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 50) | NC score | 0.927647 (rank : 38) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P05121 | Gene names | SERPINE1, PAI1, PLANH1 | |||
|
Domain Architecture |
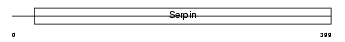
|
|||||
| Description | Plasminogen activator inhibitor 1 precursor (PAI-1) (Endothelial plasminogen activator inhibitor) (PAI). | |||||
|
HEP2_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 51) | NC score | 0.935569 (rank : 17) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P49182 | Gene names | Serpind1, Hcf2, Hcii | |||
|
Domain Architecture |

|
|||||
| Description | Heparin cofactor 2 precursor (Heparin cofactor II) (HC-II) (Protease inhibitor leuserpin 2). | |||||
|
SPB10_HUMAN
|
||||||
| θ value | 1.74391e-19 (rank : 52) | NC score | 0.921074 (rank : 57) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P48595, Q4VAX4 | Gene names | SERPINB10, PI10 | |||
|
Domain Architecture |
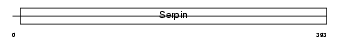
|
|||||
| Description | Serpin B10 (Bomapin) (Protease inhibitor 10). | |||||
|
CBG_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 53) | NC score | 0.926033 (rank : 44) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q06770 | Gene names | Serpina6, Cbg | |||
|
Domain Architecture |
|
|||||
| Description | Corticosteroid-binding globulin precursor (CBG) (Transcortin) (Serpin A6). | |||||
|
SPB10_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 54) | NC score | 0.919555 (rank : 61) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8K1K6 | Gene names | Serpinb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B10. | |||||
|
SPB12_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 55) | NC score | 0.918486 (rank : 63) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q96P63 | Gene names | SERPINB12 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B12. | |||||
|
A1ATR_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 56) | NC score | 0.922321 (rank : 52) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P20848 | Gene names | SERPINA2, ARGS, ATR, PIL | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1-antitrypsin-related protein precursor. | |||||
|
THBG_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 57) | NC score | 0.927450 (rank : 40) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P61939 | Gene names | Serpina7, Tbg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroxine-binding globulin precursor (T4-binding globulin) (Serpin A7). | |||||
|
SPA12_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 58) | NC score | 0.929418 (rank : 31) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8IW75 | Gene names | SERPINA12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A12 precursor (Visceral adipose-specific serpin) (Visceral adipose tissue-derived serine protease inhibitor) (Vaspin) (OL-64). | |||||
|
SPA9_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 59) | NC score | 0.926775 (rank : 41) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q86WD7, Q6UWP9, Q86WD6, Q86YP6, Q86YP7 | Gene names | SERPINA9, GCET1, SERPINA11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A9 precursor (Germinal center B-cell expressed transcript 1 protein). | |||||
|
PAI1_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 60) | NC score | 0.928075 (rank : 36) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P22777 | Gene names | Serpine1, Mr1, Pai1, Planh1 | |||
|
Domain Architecture |
|
|||||
| Description | Plasminogen activator inhibitor 1 precursor (PAI-1) (Endothelial plasminogen activator inhibitor) (PAI). | |||||
|
SPB11_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 61) | NC score | 0.916807 (rank : 66) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q96P15, Q96P13, Q96P14 | Gene names | SERPINB11 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B11. | |||||
|
SPB11_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 62) | NC score | 0.916682 (rank : 67) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9CQV3, Q91Z12 | Gene names | Serpinb11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B11. | |||||
|
SERPH_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 63) | NC score | 0.921442 (rank : 56) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P50454, P29043, Q5XPB4, Q6NSJ6, Q8IY96, Q9NP88 | Gene names | SERPINH1, CBP1, CBP2, HSP47, PIG14, SERPINH2 | |||
|
Domain Architecture |
|
|||||
| Description | Serpin H1 precursor (Collagen-binding protein) (Colligin) (47 kDa heat shock protein) (Rheumatoid arthritis-related antigen RA-A47) (Arsenic- transactivated protein 3) (AsTP3) (Proliferation-inducing gene 14 protein). | |||||
|
SERPH_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 64) | NC score | 0.920212 (rank : 59) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P19324 | Gene names | Serpinh1, Cbp1, Hsp47 | |||
|
Domain Architecture |
|
|||||
| Description | Serpin H1 precursor (Collagen-binding protein) (Colligin) (47 kDa heat shock protein) (Serine protease inhibitor J6). | |||||
|
SPB7_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 65) | NC score | 0.911332 (rank : 71) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O75635, Q3KPG4 | Gene names | SERPINB7 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B7 (Megsin) (TP55). | |||||
|
SPB6_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 66) | NC score | 0.913793 (rank : 68) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q60854 | Gene names | Serpinb6, Serpinb6a, Spi3 | |||
|
Domain Architecture |
|
|||||
| Description | Serpin B6 (Placental thrombin inhibitor) (Protease inhibitor 6) (PI- 6). | |||||
|
GDN_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 67) | NC score | 0.921610 (rank : 54) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q07235, Q921T7 | Gene names | Serpine2, Pi7, Pn1, Spi4 | |||
|
Domain Architecture |
|
|||||
| Description | Glia-derived nexin precursor (GDN) (Protease nexin I) (PN-1) (Serine protease-inhibitor 4). | |||||
|
GDN_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 68) | NC score | 0.920092 (rank : 60) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P07093 | Gene names | SERPINE2, PI7, PN1 | |||
|
Domain Architecture |
|
|||||
| Description | Glia-derived nexin precursor (GDN) (Protease nexin I) (PN-1) (Protease inhibitor 7). | |||||
|
HEP2_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 69) | NC score | 0.931512 (rank : 27) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P05546, Q6IBZ5 | Gene names | SERPIND1, HCF2 | |||
|
Domain Architecture |

|
|||||
| Description | Heparin cofactor 2 precursor (Heparin cofactor II) (HC-II) (Protease inhibitor leuserpin 2) (HLS2). | |||||
|
SPB7_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 70) | NC score | 0.913372 (rank : 69) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9D695 | Gene names | Serpinb7 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B7 (Megsin). | |||||
|
SPB13_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 71) | NC score | 0.907846 (rank : 72) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UIV8, Q9HCX1, Q9UBW1, Q9UKG0 | Gene names | SERPINB13, PI13 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B13 (Hurpin) (HaCaT UV-repressible serpin) (Protease inhibitor 13) (Headpin). | |||||
|
SPB13_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 72) | NC score | 0.912688 (rank : 70) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8CDC0 | Gene names | Serpinb13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B13. | |||||
|
SPB5_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 73) | NC score | 0.906160 (rank : 73) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P70124 | Gene names | Serpinb5, Pi5, Spi5, Spi7 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B5 precursor (Maspin) (Protease inhibitor 5). | |||||
|
SPB5_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 74) | NC score | 0.904048 (rank : 74) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P36952 | Gene names | SERPINB5, PI5 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B5 precursor (Maspin) (Protease inhibitor 5). | |||||
|
ANGT_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 75) | NC score | 0.808550 (rank : 76) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P01019, Q16358, Q16359, Q96F91 | Gene names | AGT, SERPINA8 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensinogen precursor [Contains: Angiotensin-1 (Angiotensin I) (Ang I); Angiotensin-2 (Angiotensin II) (Ang II); Angiotensin-3 (Angiotensin III) (Ang III) (Des-Asp[1]-angiotensin II)]. | |||||
|
ANGT_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 76) | NC score | 0.771542 (rank : 77) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P11859 | Gene names | Agt, Serpina8 | |||
|
Domain Architecture |
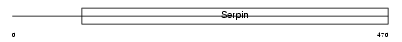
|
|||||
| Description | Angiotensinogen precursor [Contains: Angiotensin-1 (Angiotensin I) (Ang I); Angiotensin-2 (Angiotensin II) (Ang II); Angiotensin-3 (Angiotensin III) (Ang III) (Des-Asp[1]-angiotensin II)]. | |||||
|
SPA13_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 77) | NC score | 0.849718 (rank : 75) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q6UXR4 | Gene names | SERPINA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A13 precursor. | |||||
|
GP152_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 78) | NC score | 0.021920 (rank : 88) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 79) | NC score | 0.041039 (rank : 78) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 80) | NC score | 0.008219 (rank : 128) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 81) | NC score | 0.016697 (rank : 101) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 82) | NC score | 0.026834 (rank : 83) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.125558 (rank : 83) | NC score | 0.028697 (rank : 81) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 84) | NC score | 0.017309 (rank : 100) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 85) | NC score | 0.026227 (rank : 85) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.365318 (rank : 86) | NC score | 0.014675 (rank : 107) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 87) | NC score | 0.020935 (rank : 89) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 88) | NC score | 0.007226 (rank : 132) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
G3BP2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 89) | NC score | 0.017352 (rank : 99) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UN86, O60606, O75149, Q9UPA1 | Gene names | G3BP2, KIAA0660 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 90) | NC score | 0.018229 (rank : 97) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 91) | NC score | 0.028424 (rank : 82) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 92) | NC score | 0.008022 (rank : 129) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.62314 (rank : 93) | NC score | 0.018803 (rank : 94) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
THSD1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 94) | NC score | 0.024470 (rank : 86) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JM61, Q8C7Q9 | Gene names | Thsd1, Tmtsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 95) | NC score | -0.002068 (rank : 158) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 0.813845 (rank : 96) | NC score | 0.012758 (rank : 112) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
EWS_HUMAN
|
||||||
| θ value | 1.06291 (rank : 97) | NC score | 0.014653 (rank : 108) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 98) | NC score | 0.016241 (rank : 102) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
THSD1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 99) | NC score | 0.022106 (rank : 87) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 100) | NC score | 0.003715 (rank : 148) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 101) | NC score | 0.013300 (rank : 110) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 102) | NC score | 0.013252 (rank : 111) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
G3BP2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 103) | NC score | 0.015818 (rank : 104) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
GP152_MOUSE
|
||||||
| θ value | 1.38821 (rank : 104) | NC score | 0.012232 (rank : 113) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
M4K2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 105) | NC score | -0.005665 (rank : 163) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12851 | Gene names | MAP4K2, GCK, RAB8IP | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 2 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 2) (MEK kinase kinase 2) (MEKKK 2) (Germinal center kinase) (GC kinase) (Rab8-interacting protein) (B lymphocyte serine/threonine-protein kinase). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 106) | NC score | 0.026470 (rank : 84) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 107) | NC score | 0.018567 (rank : 95) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
CK024_MOUSE
|
||||||
| θ value | 1.81305 (rank : 108) | NC score | 0.029123 (rank : 80) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
F100A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 109) | NC score | 0.019933 (rank : 91) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P3B2 | Gene names | Fam100a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM100A. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 110) | NC score | 0.005373 (rank : 137) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PARM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 111) | NC score | 0.032203 (rank : 79) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PO2F1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 112) | NC score | 0.004771 (rank : 142) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 113) | NC score | 0.019038 (rank : 92) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 114) | NC score | 0.019001 (rank : 93) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 1.81305 (rank : 115) | NC score | 0.008755 (rank : 126) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 116) | NC score | -0.005442 (rank : 161) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
EWS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 117) | NC score | 0.012224 (rank : 114) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
F100A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 118) | NC score | 0.018346 (rank : 96) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TB05, Q71MF6 | Gene names | FAM100A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM100A. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 119) | NC score | 0.014940 (rank : 106) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 120) | NC score | 0.004942 (rank : 139) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
PLSL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 121) | NC score | 0.007758 (rank : 130) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
RTN4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 122) | NC score | 0.010968 (rank : 119) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 123) | NC score | -0.003034 (rank : 159) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
REPS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 124) | NC score | 0.011186 (rank : 118) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 3.0926 (rank : 125) | NC score | 0.009620 (rank : 121) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
3BP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.008823 (rank : 125) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
CD008_HUMAN
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.010702 (rank : 120) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.015690 (rank : 105) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.005625 (rank : 136) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HXA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.000059 (rank : 155) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
K0319_MOUSE
|
||||||
| θ value | 4.03905 (rank : 131) | NC score | 0.011658 (rank : 116) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5SZV5, Q80U39 | Gene names | Kiaa0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
PORIM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 132) | NC score | 0.020463 (rank : 90) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91Z22, Q3T9F6, Q3TLL2, Q3U1R6, Q8CEX4 | Gene names | Tmem123 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 133) | NC score | 0.015880 (rank : 103) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 134) | NC score | 0.011966 (rank : 115) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 135) | NC score | 0.005319 (rank : 138) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ATF5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.009068 (rank : 124) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.005947 (rank : 135) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.003129 (rank : 150) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.001927 (rank : 153) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
NFAC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.004260 (rank : 146) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
PLSL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.006726 (rank : 133) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.004874 (rank : 140) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
SON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.011301 (rank : 117) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TTLL4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.013415 (rank : 109) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
FMNL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.003107 (rank : 151) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
HXA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | -0.000074 (rank : 157) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.004715 (rank : 143) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.018023 (rank : 98) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.003940 (rank : 147) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PI51C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.004840 (rank : 141) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.004284 (rank : 145) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.006319 (rank : 134) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
WDFY3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.000333 (rank : 154) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
BOLL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.002094 (rank : 152) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q924M5, Q9D4V2 | Gene names | Boll, Boule | |||
|
Domain Architecture |
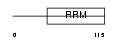
|
|||||
| Description | Protein boule-like. | |||||
|
CDCA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.008491 (rank : 127) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96FF9 | Gene names | CDCA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sororin (Cell division cycle-associated protein 5) (p35). | |||||
|
DCC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | -0.003085 (rank : 160) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
HNRL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.003162 (rank : 149) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
IRAK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | -0.005618 (rank : 162) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |
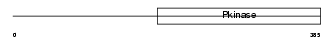
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | -0.010164 (rank : 164) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MYBPH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | -0.000026 (rank : 156) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70402 | Gene names | Mybph | |||
|
Domain Architecture |
|
|||||
| Description | Myosin-binding protein H (MyBP-H) (H-protein). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.004495 (rank : 144) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
REPS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.009399 (rank : 123) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.009437 (rank : 122) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TRML2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.007550 (rank : 131) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2LA85 | Gene names | Treml2, Tlt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 2 protein precursor (TLT-2) (Triggering receptor expressed on myeloid cells-like protein 2). | |||||
|
IC1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 164 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
IC1_HUMAN
|
||||||
| NC score | 0.990168 (rank : 2) | θ value | 2.10118e-182 (rank : 2) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P05155, Q547W3, Q59EI5, Q96FE0 | Gene names | SERPING1, C1IN, C1NH | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
A2AP_MOUSE
|
||||||
| NC score | 0.954778 (rank : 3) | θ value | 1.3722e-40 (rank : 3) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61247 | Gene names | Serpinf2, Pli | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-antiplasmin precursor (Alpha-2-plasmin inhibitor) (Alpha-2-PI) (Alpha-2-AP). | |||||
|
A2AP_HUMAN
|
||||||
| NC score | 0.954354 (rank : 4) | θ value | 2.86122e-38 (rank : 4) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P08697 | Gene names | SERPINF2, AAP, PLI | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-antiplasmin precursor (Alpha-2-plasmin inhibitor) (Alpha-2-PI) (Alpha-2-AP). | |||||
|
PEDF_MOUSE
|
||||||
| NC score | 0.949033 (rank : 5) | θ value | 3.74554e-30 (rank : 9) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P97298, O70629, O88691 | Gene names | Serpinf1, Pedf, Sdf3 | |||
|
Domain Architecture |

|
|||||
| Description | Pigment epithelium-derived factor precursor (PEDF) (Serpin-F1) (Stromal cell-derived factor 3) (SDF-3) (Caspin). | |||||
|
PEDF_HUMAN
|
||||||
| NC score | 0.948485 (rank : 6) | θ value | 1.6813e-30 (rank : 8) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P36955, Q13236, Q96CT1, Q96R01, Q9BWA4 | Gene names | SERPINF1, PEDF | |||
|
Domain Architecture |

|
|||||
| Description | Pigment epithelium-derived factor precursor (PEDF) (Serpin-F1) (EPC- 1). | |||||
|
ZPI_HUMAN
|
||||||
| NC score | 0.939904 (rank : 7) | θ value | 8.36355e-22 (rank : 39) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UK55, Q6UWX9, Q86U20 | Gene names | SERPINA10, ZPI | |||
|
Domain Architecture |

|
|||||
| Description | Protein Z-dependent protease inhibitor precursor (PZ-dependent protease inhibitor) (PZI) (Serpin A10). | |||||
|
AACT_HUMAN
|
||||||
| NC score | 0.939834 (rank : 8) | θ value | 4.42448e-31 (rank : 7) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P01011, Q13703, Q59GP9, Q6LBY8, Q6LDT7, Q6NSC9, Q8N177, Q96DW8, Q9UNU9 | Gene names | SERPINA3, AACT | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antichymotrypsin precursor (ACT) [Contains: Alpha-1- antichymotrypsin His-Pro-less]. | |||||
|
SPI2_MOUSE
|
||||||
| NC score | 0.938159 (rank : 9) | θ value | 2.12685e-25 (rank : 17) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9JK88, Q9D8Z3, Q9D955 | Gene names | Serpini2, Spi14 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin I2 precursor (Serine protease inhibitor 14). | |||||
|
SPI2_HUMAN
|
||||||
| NC score | 0.937915 (rank : 10) | θ value | 3.62785e-25 (rank : 18) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O75830 | Gene names | SERPINI2, MEPI, PI14 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin I2 precursor (Myoepithelium-derived serine protease inhibitor) (Pancpin) (Protease inhibitor 14) (TSA2004). | |||||
|
ZPI_MOUSE
|
||||||
| NC score | 0.937638 (rank : 11) | θ value | 5.42112e-21 (rank : 44) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8R121, Q8VCV8 | Gene names | Serpina10, Zpi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Z-dependent protease inhibitor precursor (PZ-dependent protease inhibitor) (PZI) (Serpin A10). | |||||
|
SPA3K_MOUSE
|
||||||
| NC score | 0.936726 (rank : 12) | θ value | 2.77131e-33 (rank : 5) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P07759, Q91W80, Q91X80 | Gene names | Serpina3k, Mcm2, Spi2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor A3K precursor (Contrapsin) (Serpin A3K). | |||||
|
SPA3C_MOUSE
|
||||||
| NC score | 0.936566 (rank : 13) | θ value | 4.14116e-29 (rank : 10) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P29621 | Gene names | Serpina3c, Klkbp | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor A3C precursor (Kallikrein-binding protein) (KBP) (Serpin A3C). | |||||
|
IPSP_HUMAN
|
||||||
| NC score | 0.936467 (rank : 14) | θ value | 1.92365e-26 (rank : 13) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P05154, Q07616, Q9UG30 | Gene names | SERPINA5, PCI, PLANH3, PROCI | |||
|
Domain Architecture |

|
|||||
| Description | Plasma serine protease inhibitor precursor (PCI) (Protein C inhibitor) (Serpin A5) (Plasminogen activator inhibitor 3) (PAI-3) (PAI3) (Acrosomal serine protease inhibitor). | |||||
|
SPA3M_MOUSE
|
||||||
| NC score | 0.936112 (rank : 15) | θ value | 1.79631e-32 (rank : 6) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q03734, Q62260 | Gene names | Serpina3m | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3M precursor (Serpin A3M). | |||||
|
SPA12_MOUSE
|
||||||
| NC score | 0.935776 (rank : 16) | θ value | 3.17815e-21 (rank : 42) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q7TMF5, Q9CQ32 | Gene names | Serpina12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A12 precursor (Visceral adipose-specific serpin) (Visceral adipose tissue-derived serine protease inhibitor) (Vaspin). | |||||
|
HEP2_MOUSE
|
||||||
| NC score | 0.935569 (rank : 17) | θ value | 1.74391e-19 (rank : 51) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P49182 | Gene names | Serpind1, Hcf2, Hcii | |||
|
Domain Architecture |

|
|||||
| Description | Heparin cofactor 2 precursor (Heparin cofactor II) (HC-II) (Protease inhibitor leuserpin 2). | |||||
|
NEUS_HUMAN
|
||||||
| NC score | 0.934766 (rank : 18) | θ value | 1.16704e-23 (rank : 27) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q99574, Q6AHZ4 | Gene names | SERPINI1, PI12 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroserpin precursor (Serpin I1) (Protease inhibitor 12). | |||||
|
SPA3G_MOUSE
|
||||||
| NC score | 0.934305 (rank : 19) | θ value | 2.51237e-26 (rank : 15) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q5I2A0, Q6PG99 | Gene names | Serpina3g, Spi2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3G (Serine protease inhibitor 2A) (Serpin A3G) (Serpin 2A). | |||||
|
SPA3F_MOUSE
|
||||||
| NC score | 0.933984 (rank : 20) | θ value | 6.61148e-27 (rank : 12) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q80X76 | Gene names | Serpina3f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3F (Serpin A3F). | |||||
|
SPA3B_MOUSE
|
||||||
| NC score | 0.933728 (rank : 21) | θ value | 1.92365e-26 (rank : 14) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8BYY9 | Gene names | Serpina3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3B precursor (Serpin A3B). | |||||
|
CBG_HUMAN
|
||||||
| NC score | 0.933318 (rank : 22) | θ value | 3.51386e-20 (rank : 47) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P08185, Q7Z2Q9 | Gene names | SERPINA6, CBG | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid-binding globulin precursor (CBG) (Transcortin) (Serpin A6). | |||||
|
IPSP_MOUSE
|
||||||
| NC score | 0.933317 (rank : 23) | θ value | 5.23862e-24 (rank : 24) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P70458 | Gene names | Serpina5, Pci | |||
|
Domain Architecture |

|
|||||
| Description | Plasma serine protease inhibitor precursor (PCI) (Protein C inhibitor) (Serpin A5) (Plasminogen activator inhibitor 3) (PAI-3) (PAI3). | |||||
|
SPA3N_MOUSE
|
||||||
| NC score | 0.932994 (rank : 24) | θ value | 1.73987e-27 (rank : 11) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q91WP6, Q62258 | Gene names | Serpina3n, Spi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3N precursor (Serpin A3N). | |||||
|
ANT3_MOUSE
|
||||||
| NC score | 0.932544 (rank : 25) | θ value | 4.43474e-23 (rank : 33) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P32261 | Gene names | Serpinc1, At3 | |||
|
Domain Architecture |

|
|||||
| Description | Antithrombin-III precursor (ATIII). | |||||
|
ANT3_HUMAN
|
||||||
| NC score | 0.931894 (rank : 26) | θ value | 8.93572e-24 (rank : 26) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P01008, P78439, P78447, Q13815, Q7KZ43 | Gene names | SERPINC1, AT3 | |||
|
Domain Architecture |

|
|||||
| Description | Antithrombin-III precursor (ATIII). | |||||
|
HEP2_HUMAN
|
||||||
| NC score | 0.931512 (rank : 27) | θ value | 2.35696e-16 (rank : 69) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P05546, Q6IBZ5 | Gene names | SERPIND1, HCF2 | |||
|
Domain Architecture |

|
|||||
| Description | Heparin cofactor 2 precursor (Heparin cofactor II) (HC-II) (Protease inhibitor leuserpin 2) (HLS2). | |||||
|
SPA9_MOUSE
|
||||||
| NC score | 0.931487 (rank : 28) | θ value | 5.23862e-24 (rank : 25) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9D7D2 | Gene names | Serpina9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A9 precursor. | |||||
|
A1AT_HUMAN
|
||||||
| NC score | 0.931315 (rank : 29) | θ value | 3.39556e-23 (rank : 32) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P01009, Q13672, Q5U0M1, Q96BF9, Q96ES1, Q9P1P0 | Gene names | SERPINA1, AAT, PI | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin precursor (Alpha-1 protease inhibitor) (Alpha-1- antiproteinase). | |||||
|
THBG_HUMAN
|
||||||
| NC score | 0.930208 (rank : 30) | θ value | 9.87957e-23 (rank : 36) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P05543 | Gene names | SERPINA7, TBG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroxine-binding globulin precursor (T4-binding globulin) (Serpin A7). | |||||
|
SPA12_HUMAN
|
||||||
| NC score | 0.929418 (rank : 31) | θ value | 4.29542e-18 (rank : 58) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8IW75 | Gene names | SERPINA12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A12 precursor (Visceral adipose-specific serpin) (Visceral adipose tissue-derived serine protease inhibitor) (Vaspin) (OL-64). | |||||
|
A1AT5_MOUSE
|
||||||
| NC score | 0.929126 (rank : 32) | θ value | 6.18819e-25 (rank : 19) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q00898 | Gene names | Serpina1e, Dom5, Spi1-5 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin 1-5 precursor (Serine protease inhibitor 1-5) (Alpha-1 protease inhibitor 5). | |||||
|
SPA3A_MOUSE
|
||||||
| NC score | 0.928727 (rank : 33) | θ value | 6.40375e-22 (rank : 38) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q6P4P1, Q9D490 | Gene names | Serpina3a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3A precursor (Serpin A3A). | |||||
|
KAIN_HUMAN
|
||||||
| NC score | 0.928508 (rank : 34) | θ value | 4.01107e-24 (rank : 23) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P29622, Q53XB5, Q86TR9, Q96BZ5 | Gene names | SERPINA4, KST, PI4 | |||
|
Domain Architecture |

|
|||||
| Description | Kallistatin precursor (Serpin A4) (Kallikrein inhibitor) (Protease inhibitor 4). | |||||
|
A1AT4_MOUSE
|
||||||
| NC score | 0.928470 (rank : 35) | θ value | 3.39556e-23 (rank : 31) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q00897 | Gene names | Serpina1d, Dom4, Spi1-4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin 1-4 precursor (Serine protease inhibitor 1-4) (Alpha-1 protease inhibitor 4). | |||||
|
PAI1_MOUSE
|
||||||
| NC score | 0.928075 (rank : 36) | θ value | 9.56915e-18 (rank : 60) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P22777 | Gene names | Serpine1, Mr1, Pai1, Planh1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen activator inhibitor 1 precursor (PAI-1) (Endothelial plasminogen activator inhibitor) (PAI). | |||||
|
A1AT1_MOUSE
|
||||||
| NC score | 0.928049 (rank : 37) | θ value | 5.79196e-23 (rank : 34) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P07758, Q80YB8, Q8JZV6, Q91XB8 | Gene names | Serpina1a, Dom1, Spi1-1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin 1-1 precursor (Serine protease inhibitor 1-1) (Alpha-1 protease inhibitor 1) (Alpha-1-antiproteinase) (AAT). | |||||
|
PAI1_HUMAN
|
||||||
| NC score | 0.927647 (rank : 38) | θ value | 1.02238e-19 (rank : 50) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P05121 | Gene names | SERPINE1, PAI1, PLANH1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen activator inhibitor 1 precursor (PAI-1) (Endothelial plasminogen activator inhibitor) (PAI). | |||||
|
NEUS_MOUSE
|
||||||
| NC score | 0.927556 (rank : 39) | θ value | 7.0802e-21 (rank : 45) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O35684, Q543F7, Q922U6 | Gene names | Serpini1, Pi12, Spi17 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroserpin precursor (Serpin I1) (Protease inhibitor 17). | |||||
|
THBG_MOUSE
|
||||||
| NC score | 0.927450 (rank : 40) | θ value | 3.28887e-18 (rank : 57) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P61939 | Gene names | Serpina7, Tbg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroxine-binding globulin precursor (T4-binding globulin) (Serpin A7). | |||||
|
SPA9_HUMAN
|
||||||
| NC score | 0.926775 (rank : 41) | θ value | 4.29542e-18 (rank : 59) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q86WD7, Q6UWP9, Q86WD6, Q86YP6, Q86YP7 | Gene names | SERPINA9, GCET1, SERPINA11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A9 precursor (Germinal center B-cell expressed transcript 1 protein). | |||||
|
SPA11_HUMAN
|
||||||
| NC score | 0.926552 (rank : 42) | θ value | 2.43343e-21 (rank : 41) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q86U17 | Gene names | SERPINA11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A11 precursor. | |||||
|
SPA11_MOUSE
|
||||||
| NC score | 0.926175 (rank : 43) | θ value | 1.86321e-21 (rank : 40) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8CIE0 | Gene names | Serpina11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A11 precursor. | |||||
|
CBG_MOUSE
|
||||||
| NC score | 0.926033 (rank : 44) | θ value | 8.65492e-19 (rank : 53) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q06770 | Gene names | Serpina6, Cbg | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid-binding globulin precursor (CBG) (Transcortin) (Serpin A6). | |||||
|
SPB3_HUMAN
|
||||||
| NC score | 0.926015 (rank : 45) | θ value | 7.30988e-26 (rank : 16) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P29508, Q96J21 | Gene names | SERPINB3, SCCA, SCCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B3 (Squamous cell carcinoma antigen 1) (SCCA-1) (Protein T4-A). | |||||
|
A1AT2_MOUSE
|
||||||
| NC score | 0.925189 (rank : 46) | θ value | 1.5242e-23 (rank : 28) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P22599, Q61283, Q80ZH5, Q8VC20 | Gene names | Serpina1b, Aat2, Dom2, Spi1-2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin 1-2 precursor (Serine protease inhibitor 1-2) (Alpha-1 protease inhibitor 2) (Alpha-1-antiproteinase) (AAT). | |||||
|
A1AT3_MOUSE
|
||||||
| NC score | 0.925038 (rank : 47) | θ value | 2.59989e-23 (rank : 30) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q00896 | Gene names | Serpina1c, Dom3, Spi1-3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin 1-3 precursor (Serine protease inhibitor 1-3) (Alpha-1 protease inhibitor 3). | |||||
|
PAI2_MOUSE
|
||||||
| NC score | 0.924875 (rank : 48) | θ value | 1.5242e-23 (rank : 29) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P12388, O35687, Q9QWP6, Q9QWP7, Q9QWP8, Q9QWP9, Q9QWQ0, Q9QWZ5, Q9QWZ6 | Gene names | Serpinb2, Pai2, Planh2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen activator inhibitor 2, macrophage (PAI-2). | |||||
|
A1AT6_MOUSE
|
||||||
| NC score | 0.924382 (rank : 49) | θ value | 9.87957e-23 (rank : 35) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P81105, Q91V74, Q91WH5, Q91XC1 | Gene names | Spi1-6, Dom6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1-antitrypsin 1-6 precursor (Serine protease inhibitor 1-6) (Alpha-1 protease inhibitor 6). | |||||
|
SPB4_HUMAN
|
||||||
| NC score | 0.924028 (rank : 50) | θ value | 2.35151e-24 (rank : 21) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P48594 | Gene names | SERPINB4, SCCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B4 (Squamous cell carcinoma antigen 2) (SCCA-2) (Leupin). | |||||
|
SPB9_HUMAN
|
||||||
| NC score | 0.923306 (rank : 51) | θ value | 7.0802e-21 (rank : 46) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P50453 | Gene names | SERPINB9, PI9 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B9 (Cytoplasmic antiproteinase 3) (CAP-3) (CAP3) (Protease inhibitor 9). | |||||
|
A1ATR_HUMAN
|
||||||
| NC score | 0.922321 (rank : 52) | θ value | 3.28887e-18 (rank : 56) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P20848 | Gene names | SERPINA2, ARGS, ATR, PIL | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin-related protein precursor. | |||||
|
SPB8_MOUSE
|
||||||
| NC score | 0.921646 (rank : 53) | θ value | 3.07116e-24 (rank : 22) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O08800 | Gene names | Serpinb8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B8. | |||||
|
GDN_MOUSE
|
||||||
| NC score | 0.921610 (rank : 54) | θ value | 1.80466e-16 (rank : 67) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q07235, Q921T7 | Gene names | Serpine2, Pi7, Pn1, Spi4 | |||
|
Domain Architecture |

|
|||||
| Description | Glia-derived nexin precursor (GDN) (Protease nexin I) (PN-1) (Serine protease-inhibitor 4). | |||||
|
ILEU_HUMAN
|
||||||
| NC score | 0.921552 (rank : 55) | θ value | 7.82807e-20 (rank : 49) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P30740, Q53FB9, Q5W0E1 | Gene names | SERPINB1, ELANH2, PI2 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte elastase inhibitor (LEI) (Serpin B1) (Monocyte/neutrophil elastase inhibitor) (M/NEI) (EI). | |||||
|
SERPH_HUMAN
|
||||||
| NC score | 0.921442 (rank : 56) | θ value | 6.20254e-17 (rank : 63) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P50454, P29043, Q5XPB4, Q6NSJ6, Q8IY96, Q9NP88 | Gene names | SERPINH1, CBP1, CBP2, HSP47, PIG14, SERPINH2 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin H1 precursor (Collagen-binding protein) (Colligin) (47 kDa heat shock protein) (Rheumatoid arthritis-related antigen RA-A47) (Arsenic- transactivated protein 3) (AsTP3) (Proliferation-inducing gene 14 protein). | |||||
|
SPB10_HUMAN
|
||||||
| NC score | 0.921074 (rank : 57) | θ value | 1.74391e-19 (rank : 52) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P48595, Q4VAX4 | Gene names | SERPINB10, PI10 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B10 (Bomapin) (Protease inhibitor 10). | |||||
|
SPB8_HUMAN
|
||||||
| NC score | 0.920331 (rank : 58) | θ value | 8.08199e-25 (rank : 20) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P50452 | Gene names | SERPINB8, PI8 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B8 (Cytoplasmic antiproteinase 2) (CAP-2) (CAP2) (Protease inhibitor 8). | |||||
|
SERPH_MOUSE
|
||||||
| NC score | 0.920212 (rank : 59) | θ value | 8.10077e-17 (rank : 64) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P19324 | Gene names | Serpinh1, Cbp1, Hsp47 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin H1 precursor (Collagen-binding protein) (Colligin) (47 kDa heat shock protein) (Serine protease inhibitor J6). | |||||
|
GDN_HUMAN
|
||||||
| NC score | 0.920092 (rank : 60) | θ value | 2.35696e-16 (rank : 68) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P07093 | Gene names | SERPINE2, PI7, PN1 | |||
|
Domain Architecture |

|
|||||
| Description | Glia-derived nexin precursor (GDN) (Protease nexin I) (PN-1) (Protease inhibitor 7). | |||||
|
SPB10_MOUSE
|
||||||
| NC score | 0.919555 (rank : 61) | θ value | 2.5182e-18 (rank : 54) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8K1K6 | Gene names | Serpinb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B10. | |||||
|
PAI2_HUMAN
|
||||||
| NC score | 0.919110 (rank : 62) | θ value | 1.6852e-22 (rank : 37) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P05120, Q96E96 | Gene names | SERPINB2, PAI2, PLANH2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen activator inhibitor 2 precursor (PAI-2) (Placental plasminogen activator inhibitor) (Monocyte Arg-serpin) (Urokinase inhibitor). | |||||
|
SPB12_HUMAN
|
||||||
| NC score | 0.918486 (rank : 63) | θ value | 2.5182e-18 (rank : 55) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q96P63 | Gene names | SERPINB12 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B12. | |||||
|
SPB12_MOUSE
|
||||||
| NC score | 0.917734 (rank : 64) | θ value | 3.51386e-20 (rank : 48) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9D7P9, Q6UKZ3 | Gene names | Serpinb12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B12. | |||||
|
SPB6_HUMAN
|
||||||
| NC score | 0.917450 (rank : 65) | θ value | 3.17815e-21 (rank : 43) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P35237, Q96J44 | Gene names | SERPINB6, PI6, PTI | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B6 (Placental thrombin inhibitor) (Cytoplasmic antiproteinase) (CAP) (Protease inhibitor 6) (PI-6). | |||||
|
SPB11_HUMAN
|
||||||
| NC score | 0.916807 (rank : 66) | θ value | 1.63225e-17 (rank : 61) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q96P15, Q96P13, Q96P14 | Gene names | SERPINB11 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B11. | |||||
|
SPB11_MOUSE
|
||||||
| NC score | 0.916682 (rank : 67) | θ value | 4.74913e-17 (rank : 62) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9CQV3, Q91Z12 | Gene names | Serpinb11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B11. | |||||
|
SPB6_MOUSE
|
||||||
| NC score | 0.913793 (rank : 68) | θ value | 1.058e-16 (rank : 66) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q60854 | Gene names | Serpinb6, Serpinb6a, Spi3 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B6 (Placental thrombin inhibitor) (Protease inhibitor 6) (PI- 6). | |||||
|
SPB7_MOUSE
|
||||||
| NC score | 0.913372 (rank : 69) | θ value | 2.35696e-16 (rank : 70) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9D695 | Gene names | Serpinb7 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B7 (Megsin). | |||||
|
SPB13_MOUSE
|
||||||
| NC score | 0.912688 (rank : 70) | θ value | 7.58209e-15 (rank : 72) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8CDC0 | Gene names | Serpinb13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B13. | |||||
|
SPB7_HUMAN
|
||||||
| NC score | 0.911332 (rank : 71) | θ value | 8.10077e-17 (rank : 65) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O75635, Q3KPG4 | Gene names | SERPINB7 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B7 (Megsin) (TP55). | |||||
|
SPB13_HUMAN
|
||||||
| NC score | 0.907846 (rank : 72) | θ value | 6.85773e-16 (rank : 71) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UIV8, Q9HCX1, Q9UBW1, Q9UKG0 | Gene names | SERPINB13, PI13 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B13 (Hurpin) (HaCaT UV-repressible serpin) (Protease inhibitor 13) (Headpin). | |||||
|
SPB5_MOUSE
|
||||||
| NC score | 0.906160 (rank : 73) | θ value | 7.84624e-12 (rank : 73) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P70124 | Gene names | Serpinb5, Pi5, Spi5, Spi7 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B5 precursor (Maspin) (Protease inhibitor 5). | |||||
|
SPB5_HUMAN
|
||||||
| NC score | 0.904048 (rank : 74) | θ value | 1.9326e-10 (rank : 74) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P36952 | Gene names | SERPINB5, PI5 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B5 precursor (Maspin) (Protease inhibitor 5). | |||||
|
SPA13_HUMAN
|
||||||
| NC score | 0.849718 (rank : 75) | θ value | 7.1131e-05 (rank : 77) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q6UXR4 | Gene names | SERPINA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A13 precursor. | |||||
|
ANGT_HUMAN
|
||||||
| NC score | 0.808550 (rank : 76) | θ value | 8.97725e-08 (rank : 75) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P01019, Q16358, Q16359, Q96F91 | Gene names | AGT, SERPINA8 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensinogen precursor [Contains: Angiotensin-1 (Angiotensin I) (Ang I); Angiotensin-2 (Angiotensin II) (Ang II); Angiotensin-3 (Angiotensin III) (Ang III) (Des-Asp[1]-angiotensin II)]. | |||||
|
ANGT_MOUSE
|
||||||
| NC score | 0.771542 (rank : 77) | θ value | 3.41135e-07 (rank : 76) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P11859 | Gene names | Agt, Serpina8 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensinogen precursor [Contains: Angiotensin-1 (Angiotensin I) (Ang I); Angiotensin-2 (Angiotensin II) (Ang II); Angiotensin-3 (Angiotensin III) (Ang III) (Des-Asp[1]-angiotensin II)]. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.041039 (rank : 78) | θ value | 0.0193708 (rank : 79) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.032203 (rank : 79) | θ value | 1.81305 (rank : 111) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.029123 (rank : 80) | θ value | 1.81305 (rank : 108) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.028697 (rank : 81) | θ value | 0.125558 (rank : 83) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.028424 (rank : 82) | θ value | 0.47712 (rank : 91) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.026834 (rank : 83) | θ value | 0.0736092 (rank : 82) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.026470 (rank : 84) | θ value | 1.38821 (rank : 106) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.026227 (rank : 85) | θ value | 0.279714 (rank : 85) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
THSD1_MOUSE
|
||||||
| NC score | 0.024470 (rank : 86) | θ value | 0.62314 (rank : 94) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JM61, Q8C7Q9 | Gene names | Thsd1, Tmtsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
THSD1_HUMAN
|
||||||
| NC score | 0.022106 (rank : 87) | θ value | 1.06291 (rank : 99) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.021920 (rank : 88) | θ value | 0.0113563 (rank : 78) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.020935 (rank : 89) | θ value | 0.365318 (rank : 87) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PORIM_MOUSE
|
||||||
| NC score | 0.020463 (rank : 90) | θ value | 4.03905 (rank : 132) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91Z22, Q3T9F6, Q3TLL2, Q3U1R6, Q8CEX4 | Gene names | Tmem123 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123). | |||||
|
F100A_MOUSE
|
||||||
| NC score | 0.019933 (rank : 91) | θ value | 1.81305 (rank : 109) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P3B2 | Gene names | Fam100a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM100A. | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.019038 (rank : 92) | θ value | 1.81305 (rank : 113) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.019001 (rank : 93) | θ value | 1.81305 (rank : 114) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.018803 (rank : 94) | θ value | 0.62314 (rank : 93) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.018567 (rank : 95) | θ value | 1.38821 (rank : 107) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
F100A_HUMAN
|
||||||
| NC score | 0.018346 (rank : 96) | θ value | 2.36792 (rank : 118) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TB05, Q71MF6 | Gene names | FAM100A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM100A. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.018229 (rank : 97) | θ value | 0.47712 (rank : 90) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.018023 (rank : 98) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
G3BP2_HUMAN
|
||||||
| NC score | 0.017352 (rank : 99) | θ value | 0.47712 (rank : 89) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UN86, O60606, O75149, Q9UPA1 | Gene names | G3BP2, KIAA0660 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.017309 (rank : 100) | θ value | 0.163984 (rank : 84) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.016697 (rank : 101) | θ value | 0.0563607 (rank : 81) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.016241 (rank : 102) | θ value | 1.06291 (rank : 98) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.015880 (rank : 103) | θ value | 4.03905 (rank : 133) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
G3BP2_MOUSE
|
||||||
| NC score | 0.015818 (rank : 104) | θ value | 1.38821 (rank : 103) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.015690 (rank : 105) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.014940 (rank : 106) | θ value | 2.36792 (rank : 119) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.014675 (rank : 107) | θ value | 0.365318 (rank : 86) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
EWS_HUMAN
|
||||||
| NC score | 0.014653 (rank : 108) | θ value | 1.06291 (rank : 97) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
TTLL4_MOUSE
|
||||||
| NC score | 0.013415 (rank : 109) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.013300 (rank : 110) | θ value | 1.38821 (rank : 101) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.013252 (rank : 111) | θ value | 1.38821 (rank : 102) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.012758 (rank : 112) | θ value | 0.813845 (rank : 96) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.012232 (rank : 113) | θ value | 1.38821 (rank : 104) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.012224 (rank : 114) | θ value | 2.36792 (rank : 117) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.011966 (rank : 115) | θ value | 4.03905 (rank : 134) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
K0319_MOUSE
|
||||||
| NC score | 0.011658 (rank : 116) | θ value | 4.03905 (rank : 131) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5SZV5, Q80U39 | Gene names | Kiaa0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.011301 (rank : 117) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
REPS1_MOUSE
|
||||||
| NC score | 0.011186 (rank : 118) | θ value | 3.0926 (rank : 124) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
RTN4_HUMAN
|
||||||
| NC score | 0.010968 (rank : 119) | θ value | 2.36792 (rank : 122) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
CD008_HUMAN
|
||||||
| NC score | 0.010702 (rank : 120) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.009620 (rank : 121) | θ value | 3.0926 (rank : 125) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.009437 (rank : 122) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
REPS1_HUMAN
|
||||||
| NC score | 0.009399 (rank : 123) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
ATF5_HUMAN
|
||||||
| NC score | 0.009068 (rank : 124) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
3BP2_MOUSE
|
||||||
| NC score | 0.008823 (rank : 125) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.008755 (rank : 126) | θ value | 1.81305 (rank : 115) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
CDCA5_HUMAN
|
||||||
| NC score | 0.008491 (rank : 127) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96FF9 | Gene names | CDCA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sororin (Cell division cycle-associated protein 5) (p35). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.008219 (rank : 128) | θ value | 0.0330416 (rank : 80) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.008022 (rank : 129) | θ value | 0.62314 (rank : 92) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
PLSL_MOUSE
|
||||||
| NC score | 0.007758 (rank : 130) | θ value | 2.36792 (rank : 121) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
TRML2_MOUSE
|
||||||
| NC score | 0.007550 (rank : 131) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2LA85 | Gene names | Treml2, Tlt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 2 protein precursor (TLT-2) (Triggering receptor expressed on myeloid cells-like protein 2). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.007226 (rank : 132) | θ value | 0.47712 (rank : 88) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
PLSL_HUMAN
|
||||||
| NC score | 0.006726 (rank : 133) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.006319 (rank : 134) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.005947 (rank : 135) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.005625 (rank : 136) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.005373 (rank : 137) | θ value | 1.81305 (rank : 110) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.005319 (rank : 138) | θ value | 4.03905 (rank : 135) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.004942 (rank : 139) | θ value | 2.36792 (rank : 120) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.004874 (rank : 140) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
PI51C_HUMAN
|
||||||
| NC score | 0.004840 (rank : 141) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
PO2F1_HUMAN
|
||||||
| NC score | 0.004771 (rank : 142) | θ value | 1.81305 (rank : 112) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.004715 (rank : 143) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.004495 (rank : 144) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.004284 (rank : 145) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
NFAC3_MOUSE
|
||||||
| NC score | 0.004260 (rank : 146) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.003940 (rank : 147) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.003715 (rank : 148) | θ value | 1.38821 (rank : 100) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
HNRL1_MOUSE
|
||||||
| NC score | 0.003162 (rank : 149) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.003129 (rank : 150) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
FMNL_MOUSE
|
||||||
| NC score | 0.003107 (rank : 151) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
BOLL_MOUSE
|
||||||
| NC score | 0.002094 (rank : 152) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q924M5, Q9D4V2 | Gene names | Boll, Boule | |||
|
Domain Architecture |

|
|||||
| Description | Protein boule-like. | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.001927 (rank : 153) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
WDFY3_HUMAN
|
||||||
| NC score | 0.000333 (rank : 154) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
HXA3_HUMAN
|
||||||
| NC score | 0.000059 (rank : 155) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
MYBPH_MOUSE
|
||||||
| NC score | -0.000026 (rank : 156) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70402 | Gene names | Mybph | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-binding protein H (MyBP-H) (H-protein). | |||||
|
HXA3_MOUSE
|
||||||
| NC score | -0.000074 (rank : 157) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | -0.002068 (rank : 158) | θ value | 0.62314 (rank : 95) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | -0.003034 (rank : 159) | θ value | 3.0926 (rank : 123) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
DCC_HUMAN
|
||||||
| NC score | -0.003085 (rank : 160) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | -0.005442 (rank : 161) | θ value | 2.36792 (rank : 116) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
IRAK1_MOUSE
|
||||||
| NC score | -0.005618 (rank : 162) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
M4K2_HUMAN
|
||||||
| NC score | -0.005665 (rank : 163) | θ value | 1.38821 (rank : 105) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12851 | Gene names | MAP4K2, GCK, RAB8IP | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 2 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 2) (MEK kinase kinase 2) (MEKKK 2) (Germinal center kinase) (GC kinase) (Rab8-interacting protein) (B lymphocyte serine/threonine-protein kinase). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | -0.010164 (rank : 164) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||