Please be patient as the page loads
|
K0319_MOUSE
|
||||||
| SwissProt Accessions | Q5SZV5, Q80U39 | Gene names | Kiaa0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K0319_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.931523 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5VV43, Q9UJC8, Q9Y4G7 | Gene names | KIAA0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
K0319_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q5SZV5, Q80U39 | Gene names | Kiaa0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
LRP11_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 3) | NC score | 0.453807 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
LRP11_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 4) | NC score | 0.450867 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CB67, Q8C7Y7 | Gene names | Lrp11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
ABI3_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 5) | NC score | 0.083869 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYZ1, Q6PE63, Q9D7S4 | Gene names | Abi3, Nesh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 6) | NC score | 0.074791 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 7) | NC score | 0.044342 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ROBO2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 8) | NC score | 0.056745 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCK4, O43608 | Gene names | ROBO2, KIAA1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
TENN_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 9) | NC score | 0.051969 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80Z71 | Gene names | Tnn | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
SIGL8_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 10) | NC score | 0.048919 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYZ4, Q7Z728 | Gene names | SIGLEC8, SAF2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 8 precursor (Siglec-8) (Sialoadhesin family member 2) (SAF-2) (CDw329 antigen). | |||||
|
NFASC_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.053792 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q810U3 | Gene names | Nfasc | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
PCDH9_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 12) | NC score | 0.046073 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
CNTN5_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.059236 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94779, O94780 | Gene names | CNTN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2) (hNB-2). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.032694 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
I12R1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.085202 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42701 | Gene names | IL12RB1, IL12R, IL12RB | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-12 receptor beta-1 chain precursor (IL-12R-beta1) (Interleukin-12 receptor beta) (IL-12 receptor beta component) (IL- 12RB1) (CD212 antigen). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.068676 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.048433 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.056562 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
CNTN5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.054062 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P68500 | Gene names | Cntn5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2). | |||||
|
FA8_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.028602 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.049142 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.049672 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
PCDG7_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.036011 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5G6, Q9Y5D0 | Gene names | PCDHGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A7 precursor (PCDH-gamma-A7). | |||||
|
PCDGA_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.036245 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y5H3, Q9Y5E0 | Gene names | PCDHGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A10 precursor (PCDH-gamma-A10). | |||||
|
PCDGC_HUMAN
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.035911 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60330, O15100, Q6UW70, Q9Y5D7 | Gene names | PCDHGA12, CDH21, FIB3, KIAA0588 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A12 precursor (PCDH-gamma-A12) (Cadherin-21) (Fibroblast cadherin 3). | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.071317 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
ABI3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.066760 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2A4, Q9H0P6 | Gene names | ABI3, NESH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.035505 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
CT075_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.038957 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WUT4, Q5JWV6, Q9H419 | Gene names | C20orf75 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 precursor. | |||||
|
SPIT1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.133284 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.064534 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ROBO2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.051703 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
SDC3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.065141 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64519, Q3UDD9, Q6ZQA4, Q7TQD4 | Gene names | Sdc3, Kiaa0468 | |||
|
Domain Architecture |
|
|||||
| Description | Syndecan-3 precursor (SYND3). | |||||
|
CN037_MOUSE
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.053820 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.069367 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.052881 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.068333 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.041725 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
FATH_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.041367 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.045077 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
OS9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.054391 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
ARHG7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.026992 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
ARHGF_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.034225 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.047393 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
RELN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.061087 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |
|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
RELN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.061319 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60841, Q9CUA6 | Gene names | Reln, Rl | |||
|
Domain Architecture |
|
|||||
| Description | Reelin precursor (EC 3.4.21.-) (Reeler protein). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.026129 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.045327 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.081184 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.051377 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
VRK3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.026232 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IV63, Q9P2V8 | Gene names | VRK3 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein kinase VRK3 (EC 2.7.11.1) (Vaccinia-related kinase 3). | |||||
|
ARHG7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.027186 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ES28, O08757, Q9ES27 | Gene names | Arhgef7, Pak3bp | |||
|
Domain Architecture |
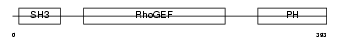
|
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (p85SPR). | |||||
|
FREM3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.031815 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5H8B9 | Gene names | Frem3, Nv2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 3 precursor (NV domain- containing protein 2). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.050712 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.029219 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
CENG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.015542 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.047200 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
NU214_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.052451 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PCD16_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.039860 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
PCDGH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.031589 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y5G0, Q9Y5C6 | Gene names | PCDHGB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B5 precursor (PCDH-gamma-B5). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.049976 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.032279 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
TNR8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.037763 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |
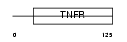
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
A1BG_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.029048 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04217, Q8IYJ6, Q96P39 | Gene names | A1BG | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1B-glycoprotein precursor (Alpha-1-B glycoprotein). | |||||
|
ADA15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.016664 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13444, Q13493, Q96C78 | Gene names | ADAM15, MDC15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 15) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein 15) (MDC-15) (Metalloprotease RGD disintegrin protein) (Metargidin). | |||||
|
COCA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.033434 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.012682 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
DFFA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.037257 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54786, O54787 | Gene names | Dffa, Icad | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA fragmentation factor subunit alpha (DNA fragmentation factor 45 kDa subunit) (DFF-45) (Inhibitor of CAD) (ICAD). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.047300 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
IRAK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.006240 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |
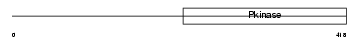
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
LIPA3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.012997 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
MYC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.036520 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.008552 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.030355 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SPIT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.117088 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.038049 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
MAGG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.021786 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |
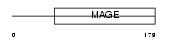
|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
MEPE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.033974 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |
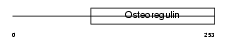
|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
PCDB5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.030348 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5E4, Q9UFU9 | Gene names | PCDHB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 5 precursor (PCDH-beta5). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.034662 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.040671 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.012785 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SDK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.043429 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6V4S5, Q3TTK1, Q5U5W7, Q6ZPP2 | Gene names | Sdk2, Kiaa1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
SIX5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.020254 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70178 | Gene names | Six5, Dmahp | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX5 (DM locus-associated homeodomain protein homolog). | |||||
|
SPT22_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.029956 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SV06 | Gene names | Spata22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 22. | |||||
|
AIM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.023754 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
CD68_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.034262 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P34810, Q96BI7 | Gene names | CD68 | |||
|
Domain Architecture |
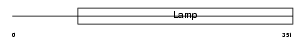
|
|||||
| Description | Macrosialin precursor (GP110) (CD68 antigen). | |||||
|
IC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.011658 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
ITB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.021852 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.072013 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.056395 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.003668 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
PCDB3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.037894 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5E6 | Gene names | PCDHB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 3 precursor (PCDH-beta3). | |||||
|
PCDG5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.032525 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5G8, Q9Y5D2 | Gene names | PCDHGA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A5 precursor (PCDH-gamma-A5). | |||||
|
PJA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.016914 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
PODXL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.048984 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R0M4, Q9ESZ1 | Gene names | Podxl, Pclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
ADA19_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.011761 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35674 | Gene names | Adam19, Mltnb | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta). | |||||
|
AP180_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.029709 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
CEAM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.034769 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P31809 | Gene names | Ceacam1, Bgp, Bgp1, Bgpd | |||
|
Domain Architecture |
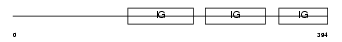
|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 1 precursor (Biliary glycoprotein 1) (BGP-1) (Murine hepatitis virus receptor) (MHV-R) (Biliary glycoprotein D). | |||||
|
LYST_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.009476 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.040628 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
PF21B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.026625 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PLCB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.009135 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.023402 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.055880 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SUPT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.029319 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75486, O76066 | Gene names | SUPT3H, SPT3 | |||
|
Domain Architecture |
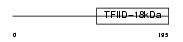
|
|||||
| Description | Transcription initiation protein SPT3 homolog (SPT3-like protein). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.027307 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.042154 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TENN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.040910 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQP3, Q5R360 | Gene names | TNN | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
TERT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.021464 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70372, O35432, Q9JK99 | Gene names | Tert | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomerase reverse transcriptase (EC 2.7.7.49) (Telomerase catalytic subunit). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.006334 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.014735 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.026032 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CSF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.025537 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07141, Q8R3C8 | Gene names | Csf1, Csfm | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF). | |||||
|
FSTL5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.035768 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.054114 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.025419 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.037953 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
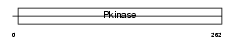
NEK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.002991 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
NOL4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.014358 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94818, Q9BWF1 | Gene names | NOL4, NOLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 4 (Nucleolar-localized protein). | |||||
|
ROBO3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.045616 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
SHD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.009863 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96IW2, Q96NC2 | Gene names | SHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein D. | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.024804 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
FUT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.008080 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21217, Q99448, Q99449 | Gene names | FUT3, FT3B, LE | |||
|
Domain Architecture |

|
|||||
| Description | Galactoside 3(4)-L-fucosyltransferase (EC 2.4.1.65) (Blood group Lewis alpha-4-fucosyltransferase) (Lewis FT) (Fucosyltransferase 3) (FUCT- III). | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.009134 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.024410 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.022015 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
ODO2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.013241 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.002881 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
SDK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.041656 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3UH53, Q3UFD1, Q6PAL2, Q6V3A4, Q8BMC2, Q8BZI1 | Gene names | Sdk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.053563 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.008216 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.005243 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
ZN143_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | -0.002378 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 718 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52747, O75559, Q8WUK9 | Gene names | ZNF143, SBF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 143 (SPH-binding factor). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.051598 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
K0319_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q5SZV5, Q80U39 | Gene names | Kiaa0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
K0319_HUMAN
|
||||||
| NC score | 0.931523 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5VV43, Q9UJC8, Q9Y4G7 | Gene names | KIAA0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
LRP11_HUMAN
|
||||||
| NC score | 0.453807 (rank : 3) | θ value | 1.38178e-16 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
LRP11_MOUSE
|
||||||
| NC score | 0.450867 (rank : 4) | θ value | 4.02038e-16 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CB67, Q8C7Y7 | Gene names | Lrp11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
SPIT1_HUMAN
|
||||||
| NC score | 0.133284 (rank : 5) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SPIT1_MOUSE
|
||||||
| NC score | 0.117088 (rank : 6) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
I12R1_HUMAN
|
||||||
| NC score | 0.085202 (rank : 7) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42701 | Gene names | IL12RB1, IL12R, IL12RB | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-12 receptor beta-1 chain precursor (IL-12R-beta1) (Interleukin-12 receptor beta) (IL-12 receptor beta component) (IL- 12RB1) (CD212 antigen). | |||||
|
ABI3_MOUSE
|
||||||
| NC score | 0.083869 (rank : 8) | θ value | 0.00228821 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYZ1, Q6PE63, Q9D7S4 | Gene names | Abi3, Nesh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.081184 (rank : 9) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.074791 (rank : 10) | θ value | 0.00228821 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.072013 (rank : 11) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.071317 (rank : 12) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.069367 (rank : 13) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.068676 (rank : 14) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.068333 (rank : 15) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
ABI3_HUMAN
|
||||||
| NC score | 0.066760 (rank : 16) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2A4, Q9H0P6 | Gene names | ABI3, NESH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
SDC3_MOUSE
|
||||||
| NC score | 0.065141 (rank : 17) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64519, Q3UDD9, Q6ZQA4, Q7TQD4 | Gene names | Sdc3, Kiaa0468 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-3 precursor (SYND3). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.064534 (rank : 18) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RELN_MOUSE
|
||||||
| NC score | 0.061319 (rank : 19) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60841, Q9CUA6 | Gene names | Reln, Rl | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-) (Reeler protein). | |||||
|
RELN_HUMAN
|
||||||
| NC score | 0.061087 (rank : 20) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
CNTN5_HUMAN
|
||||||
| NC score | 0.059236 (rank : 21) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94779, O94780 | Gene names | CNTN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2) (hNB-2). | |||||
|
ROBO2_HUMAN
|
||||||
| NC score | 0.056745 (rank : 22) | θ value | 0.00509761 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCK4, O43608 | Gene names | ROBO2, KIAA1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.056562 (rank : 23) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.056395 (rank : 24) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.055880 (rank : 25) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
OS9_HUMAN
|
||||||
| NC score | 0.054391 (rank : 26) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.054114 (rank : 27) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
CNTN5_MOUSE
|
||||||
| NC score | 0.054062 (rank : 28) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P68500 | Gene names | Cntn5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2). | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.053820 (rank : 29) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
NFASC_MOUSE
|
||||||
| NC score | 0.053792 (rank : 30) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q810U3 | Gene names | Nfasc | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.053563 (rank : 31) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.052881 (rank : 32) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.052451 (rank : 33) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
TENN_MOUSE
|
||||||
| NC score | 0.051969 (rank : 34) | θ value | 0.00665767 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80Z71 | Gene names | Tnn | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
ROBO2_MOUSE
|
||||||
| NC score | 0.051703 (rank : 35) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.051598 (rank : 36) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.051377 (rank : 37) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.050712 (rank : 38) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.049976 (rank : 39) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.049672 (rank : 40) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.049142 (rank : 41) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
PODXL_MOUSE
|
||||||
| NC score | 0.048984 (rank : 42) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R0M4, Q9ESZ1 | Gene names | Podxl, Pclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
SIGL8_HUMAN
|
||||||
| NC score | 0.048919 (rank : 43) | θ value | 0.0113563 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYZ4, Q7Z728 | Gene names | SIGLEC8, SAF2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 8 precursor (Siglec-8) (Sialoadhesin family member 2) (SAF-2) (CDw329 antigen). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.048433 (rank : 44) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.047393 (rank : 45) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.047300 (rank : 46) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.047200 (rank : 47) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
PCDH9_HUMAN
|
||||||
| NC score | 0.046073 (rank : 48) | θ value | 0.0193708 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
ROBO3_HUMAN
|
||||||
| NC score | 0.045616 (rank : 49) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.045327 (rank : 50) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.045077 (rank : 51) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.044342 (rank : 52) | θ value | 0.00298849 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
SDK2_MOUSE
|
||||||
| NC score | 0.043429 (rank : 53) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6V4S5, Q3TTK1, Q5U5W7, Q6ZPP2 | Gene names | Sdk2, Kiaa1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.042154 (rank : 54) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.041725 (rank : 55) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SDK1_MOUSE
|
||||||
| NC score | 0.041656 (rank : 56) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3UH53, Q3UFD1, Q6PAL2, Q6V3A4, Q8BMC2, Q8BZI1 | Gene names | Sdk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
FATH_HUMAN
|
||||||
| NC score | 0.041367 (rank : 57) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||
|
TENN_HUMAN
|
||||||
| NC score | 0.040910 (rank : 58) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQP3, Q5R360 | Gene names | TNN | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.040671 (rank : 59) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.040628 (rank : 60) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
PCD16_HUMAN
|
||||||
| NC score | 0.039860 (rank : 61) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
CT075_HUMAN
|
||||||
| NC score | 0.038957 (rank : 62) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WUT4, Q5JWV6, Q9H419 | Gene names | C20orf75 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 precursor. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.038049 (rank : 63) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.037953 (rank : 64) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PCDB3_HUMAN
|
||||||
| NC score | 0.037894 (rank : 65) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5E6 | Gene names | PCDHB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 3 precursor (PCDH-beta3). | |||||
|
TNR8_MOUSE
|
||||||
| NC score | 0.037763 (rank : 66) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
DFFA_MOUSE
|
||||||
| NC score | 0.037257 (rank : 67) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54786, O54787 | Gene names | Dffa, Icad | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA fragmentation factor subunit alpha (DNA fragmentation factor 45 kDa subunit) (DFF-45) (Inhibitor of CAD) (ICAD). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.036520 (rank : 68) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
PCDGA_HUMAN
|
||||||
| NC score | 0.036245 (rank : 69) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y5H3, Q9Y5E0 | Gene names | PCDHGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A10 precursor (PCDH-gamma-A10). | |||||
|
PCDG7_HUMAN
|
||||||
| NC score | 0.036011 (rank : 70) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5G6, Q9Y5D0 | Gene names | PCDHGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A7 precursor (PCDH-gamma-A7). | |||||
|
PCDGC_HUMAN
|
||||||
| NC score | 0.035911 (rank : 71) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60330, O15100, Q6UW70, Q9Y5D7 | Gene names | PCDHGA12, CDH21, FIB3, KIAA0588 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A12 precursor (PCDH-gamma-A12) (Cadherin-21) (Fibroblast cadherin 3). | |||||
|
FSTL5_MOUSE
|
||||||
| NC score | 0.035768 (rank : 72) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.035505 (rank : 73) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
CEAM1_MOUSE
|
||||||
| NC score | 0.034769 (rank : 74) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P31809 | Gene names | Ceacam1, Bgp, Bgp1, Bgpd | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 1 precursor (Biliary glycoprotein 1) (BGP-1) (Murine hepatitis virus receptor) (MHV-R) (Biliary glycoprotein D). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.034662 (rank : 75) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
CD68_HUMAN
|
||||||
| NC score | 0.034262 (rank : 76) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P34810, Q96BI7 | Gene names | CD68 | |||
|
Domain Architecture |

|
|||||
| Description | Macrosialin precursor (GP110) (CD68 antigen). | |||||
|
ARHGF_HUMAN
|
||||||
| NC score | 0.034225 (rank : 77) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
MEPE_HUMAN
|
||||||
| NC score | 0.033974 (rank : 78) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |

|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.033434 (rank : 79) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.032694 (rank : 80) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
PCDG5_HUMAN
|
||||||
| NC score | 0.032525 (rank : 81) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5G8, Q9Y5D2 | Gene names | PCDHGA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A5 precursor (PCDH-gamma-A5). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.032279 (rank : 82) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
FREM3_MOUSE
|
||||||
| NC score | 0.031815 (rank : 83) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5H8B9 | Gene names | Frem3, Nv2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 3 precursor (NV domain- containing protein 2). | |||||
|
PCDGH_HUMAN
|
||||||
| NC score | 0.031589 (rank : 84) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y5G0, Q9Y5C6 | Gene names | PCDHGB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B5 precursor (PCDH-gamma-B5). | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.030355 (rank : 85) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
PCDB5_HUMAN
|
||||||
| NC score | 0.030348 (rank : 86) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5E4, Q9UFU9 | Gene names | PCDHB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 5 precursor (PCDH-beta5). | |||||
|
SPT22_MOUSE
|
||||||
| NC score | 0.029956 (rank : 87) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SV06 | Gene names | Spata22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 22. | |||||
|
AP180_HUMAN
|
||||||
| NC score | 0.029709 (rank : 88) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
SUPT3_HUMAN
|
||||||
| NC score | 0.029319 (rank : 89) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75486, O76066 | Gene names | SUPT3H, SPT3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation protein SPT3 homolog (SPT3-like protein). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.029219 (rank : 90) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
A1BG_HUMAN
|
||||||
| NC score | 0.029048 (rank : 91) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04217, Q8IYJ6, Q96P39 | Gene names | A1BG | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1B-glycoprotein precursor (Alpha-1-B glycoprotein). | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.028602 (rank : 92) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.027307 (rank : 93) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
ARHG7_MOUSE
|
||||||
| NC score | 0.027186 (rank : 94) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ES28, O08757, Q9ES27 | Gene names | Arhgef7, Pak3bp | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (p85SPR). | |||||
|
ARHG7_HUMAN
|
||||||
| NC score | 0.026992 (rank : 95) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.026625 (rank : 96) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
VRK3_HUMAN
|
||||||
| NC score | 0.026232 (rank : 97) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IV63, Q9P2V8 | Gene names | VRK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase VRK3 (EC 2.7.11.1) (Vaccinia-related kinase 3). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.026129 (rank : 98) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.026032 (rank : 99) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CSF1_MOUSE
|
||||||
| NC score | 0.025537 (rank : 100) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07141, Q8R3C8 | Gene names | Csf1, Csfm | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.025419 (rank : 101) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.024804 (rank : 102) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.024410 (rank : 103) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
AIM1_HUMAN
|
||||||
| NC score | 0.023754 (rank : 104) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.023402 (rank : 105) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.022015 (rank : 106) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
ITB2_MOUSE
|
||||||
| NC score | 0.021852 (rank : 107) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
MAGG1_MOUSE
|
||||||
| NC score | 0.021786 (rank : 108) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
TERT_MOUSE
|
||||||
| NC score | 0.021464 (rank : 109) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70372, O35432, Q9JK99 | Gene names | Tert | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomerase reverse transcriptase (EC 2.7.7.49) (Telomerase catalytic subunit). | |||||
|
SIX5_MOUSE
|
||||||
| NC score | 0.020254 (rank : 110) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70178 | Gene names | Six5, Dmahp | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX5 (DM locus-associated homeodomain protein homolog). | |||||
|
PJA1_MOUSE
|
||||||
| NC score | 0.016914 (rank : 111) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
ADA15_HUMAN
|
||||||
| NC score | 0.016664 (rank : 112) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13444, Q13493, Q96C78 | Gene names | ADAM15, MDC15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 15) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein 15) (MDC-15) (Metalloprotease RGD disintegrin protein) (Metargidin). | |||||
|
CENG1_HUMAN
|
||||||
| NC score | 0.015542 (rank : 113) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.014735 (rank : 114) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
NOL4_HUMAN
|
||||||
| NC score | 0.014358 (rank : 115) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94818, Q9BWF1 | Gene names | NOL4, NOLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 4 (Nucleolar-localized protein). | |||||
|
ODO2_MOUSE
|
||||||
| NC score | 0.013241 (rank : 116) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
LIPA3_MOUSE
|
||||||
| NC score | 0.012997 (rank : 117) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.012785 (rank : 118) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.012682 (rank : 119) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
ADA19_MOUSE
|
||||||
| NC score | 0.011761 (rank : 120) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35674 | Gene names | Adam19, Mltnb | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta). | |||||
|
IC1_MOUSE
|
||||||
| NC score | 0.011658 (rank : 121) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
SHD_HUMAN
|
||||||
| NC score | 0.009863 (rank : 122) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96IW2, Q96NC2 | Gene names | SHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein D. | |||||
|
LYST_HUMAN
|
||||||
| NC score | 0.009476 (rank : 123) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
PLCB2_HUMAN
|
||||||
| NC score | 0.009135 (rank : 124) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.009134 (rank : 125) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.008552 (rank : 126) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.008216 (rank : 127) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
FUT3_HUMAN
|
||||||
| NC score | 0.008080 (rank : 128) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21217, Q99448, Q99449 | Gene names | FUT3, FT3B, LE | |||
|
Domain Architecture |

|
|||||
| Description | Galactoside 3(4)-L-fucosyltransferase (EC 2.4.1.65) (Blood group Lewis alpha-4-fucosyltransferase) (Lewis FT) (Fucosyltransferase 3) (FUCT- III). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.006334 (rank : 129) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
IRAK1_HUMAN
|
||||||
| NC score | 0.006240 (rank : 130) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.005243 (rank : 131) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.003668 (rank : 132) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
NEK1_HUMAN
|
||||||
| NC score | 0.002991 (rank : 133) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.002881 (rank : 134) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
ZN143_HUMAN
|
||||||
| NC score | -0.002378 (rank : 135) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 718 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52747, O75559, Q8WUK9 | Gene names | ZNF143, SBF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 143 (SPH-binding factor). | |||||