Please be patient as the page loads
|
ZCHC5_HUMAN
|
||||||
| SwissProt Accessions | Q8N8U3 | Gene names | ZCCHC5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCHC domain-containing protein 5. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZCHC5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N8U3 | Gene names | ZCCHC5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCHC domain-containing protein 5. | |||||
|
LDOC1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 2) | NC score | 0.426285 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95751 | Gene names | LDOC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LDOC1 (Leucine zipper protein down-regulated in cancer cells). | |||||
|
LDOC1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 3) | NC score | 0.390549 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPY9 | Gene names | Ldoc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LDOC1. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 4) | NC score | 0.126507 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 5) | NC score | 0.087226 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 6) | NC score | 0.111401 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 7) | NC score | 0.124350 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 8) | NC score | 0.127588 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 9) | NC score | 0.109624 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.118792 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.111851 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.116540 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
K1210_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.153076 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.116179 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.077913 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
NRBF2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.109120 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCQ3, Q9DCG3 | Gene names | Nrbf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.053223 (rank : 140) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.133362 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.095910 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.119720 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
CD248_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.042952 (rank : 161) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
ES8L2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.033928 (rank : 170) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H6S3, Q53GM8, Q8WYW7, Q96K06, Q9H6K9 | Gene names | EPS8L2, EPS8R2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.020712 (rank : 186) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
LTB1L_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.027174 (rank : 180) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.025511 (rank : 182) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
TRM6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.064086 (rank : 87) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJA5, Q76P92, Q9BQV5, Q9ULR7 | Gene names | TRM6, KIAA1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.024999 (rank : 183) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
ATS12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.019498 (rank : 187) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.028793 (rank : 178) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.066749 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
IRF7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.034093 (rank : 168) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70434 | Gene names | Irf7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.017668 (rank : 190) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
NRBF2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.090924 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96F24, Q86UR2, Q96NP6, Q9H0S9, Q9H2I2 | Gene names | NRBF2, COPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2) (Comodulator of PPAR and RXR). | |||||
|
MA1B1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.034796 (rank : 166) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKM7, Q5VSG3, Q9BRS9, Q9Y5K7 | Gene names | MAN1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (EC 3.2.1.113) (ER alpha-1,2-mannosidase) (Mannosidase alpha class 1B member 1) (Man9GlcNAc2-specific-processing alpha-mannosidase). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.045230 (rank : 160) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.078470 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PREP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.042523 (rank : 162) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K411, Q3THC4, Q3TJI1, Q3TPP0, Q3URQ8, Q4KUG2, Q4KUG3, Q6ZPX6, Q922N1, Q9CV63 | Gene names | Pitrm1, Kiaa1104, Ntup1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (Pitrilysin metalloproteinase 1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.059626 (rank : 100) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
REC8L_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.054574 (rank : 125) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5S7, Q3UIN9, Q9JK52 | Gene names | Rec8L1, Rec8 | |||
|
Domain Architecture |

|
|||||
| Description | Meiotic recombination protein REC8-like 1 (Cohesin Rec8p). | |||||
|
CO039_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.077872 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
GAK18_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.018015 (rank : 188) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.076097 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.069924 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.017868 (rank : 189) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
FOSL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.033518 (rank : 172) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.075130 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.097324 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
PO2F2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.014970 (rank : 192) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00196, Q00197, Q00198, Q00199, Q00200, Q00201, Q05882, Q61995, Q61996, Q64245 | Gene names | Pou2f2, Oct2, Otf2 | |||
|
Domain Architecture |
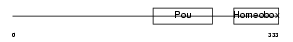
|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.053455 (rank : 138) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
CNBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.028116 (rank : 179) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.061254 (rank : 96) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.065158 (rank : 80) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.037124 (rank : 163) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SNX21_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.034910 (rank : 165) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969T3, Q5JZH6, Q9BR16 | Gene names | SNX21, C20orf161, SNXL | |||
|
Domain Architecture |
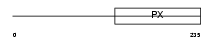
|
|||||
| Description | Sorting nexin-21 (Sorting nexin L) (SNX-L). | |||||
|
SRMS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.000750 (rank : 197) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62270, Q62360, Q923M5 | Gene names | Srms, Srm | |||
|
Domain Architecture |
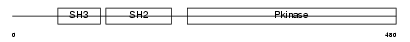
|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2) (PTK70). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.034155 (rank : 167) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.014477 (rank : 193) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CMGA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.036686 (rank : 164) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
FOSB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.033988 (rank : 169) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOSB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.033632 (rank : 171) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
HSF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.028875 (rank : 177) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
ITA5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.009302 (rank : 195) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11688 | Gene names | Itga5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
MCM3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.022069 (rank : 185) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.086592 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
ZCHC6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.031278 (rank : 173) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZCHC6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.029771 (rank : 174) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZDHC9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.013050 (rank : 194) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y397, Q59EK4, Q5JSW5, Q8WWS7, Q9BPY4, Q9NSP0, Q9NVL0, Q9NVR6 | Gene names | ZDHHC9, ZNF379 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9) (Zinc finger protein 379). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.065351 (rank : 79) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
BAI2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.009235 (rank : 196) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGM1, Q3TYC8, Q3UN11, Q3UNE2, Q6PGN0 | Gene names | Bai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
CF057_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.029738 (rank : 175) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VUM1 | Gene names | C6orf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0369 protein C6orf57 precursor. | |||||
|
CU030_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.029486 (rank : 176) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UFM2 | Gene names | C21orf30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C21orf30. | |||||
|
IZUM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.022866 (rank : 184) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9J7 | Gene names | Izumo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Izumo sperm-egg fusion protein 1 precursor (Sperm-specific protein izumo). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.017538 (rank : 191) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
SHD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.026953 (rank : 181) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96IW2, Q96NC2 | Gene names | SHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein D. | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.056802 (rank : 121) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
AFF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.068083 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.051483 (rank : 149) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.051578 (rank : 147) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
ALEX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.078375 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ALEX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.077448 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.057560 (rank : 116) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.053964 (rank : 131) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
BAG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.057558 (rank : 117) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.110457 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.100461 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.064823 (rank : 84) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.060217 (rank : 98) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.053980 (rank : 130) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.057167 (rank : 119) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
BSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.053526 (rank : 135) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.053488 (rank : 137) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.050467 (rank : 156) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.059596 (rank : 101) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.065025 (rank : 81) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CEND_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.058612 (rank : 110) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N111, Q9NYM6 | Gene names | CEND1, BM88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle exit and neuronal differentiation protein 1 (BM88 antigen). | |||||
|
CEND_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.054696 (rank : 123) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKC6, Q9DBA0 | Gene names | Cend1, Bm88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle exit and neuronal differentiation protein 1 (BM88 antigen). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.070765 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.091464 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.062473 (rank : 92) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.159945 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.078219 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
DGKK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.057141 (rank : 120) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.059525 (rank : 102) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.060394 (rank : 97) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
ENL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.079089 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
EP300_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.054319 (rank : 128) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.051171 (rank : 154) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.064826 (rank : 83) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.059241 (rank : 106) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GNAS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.051073 (rank : 155) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.089759 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
HRX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.050358 (rank : 157) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
K1802_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.052932 (rank : 144) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KI67_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.058614 (rank : 109) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.072159 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
LR37B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.059031 (rank : 108) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.057481 (rank : 118) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.073857 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.068626 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.068786 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.109418 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.061270 (rank : 95) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.065773 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.068553 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MINT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.063933 (rank : 88) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.058303 (rank : 111) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.062600 (rank : 90) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.062253 (rank : 93) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.103613 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.051265 (rank : 151) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NELFA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.053370 (rank : 139) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.068162 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.085220 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.103413 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.063098 (rank : 89) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.070442 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.053496 (rank : 136) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.051376 (rank : 150) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.053603 (rank : 133) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.055601 (rank : 122) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PHAR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.052533 (rank : 146) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
PO121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.058237 (rank : 112) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PO121_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.054061 (rank : 129) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.064861 (rank : 82) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRAX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.054660 (rank : 124) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.051571 (rank : 148) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.054453 (rank : 126) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.105449 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.104322 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.081440 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.076384 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.086458 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.057565 (rank : 115) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
PS1C2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.059265 (rank : 105) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.074495 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RERE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.059372 (rank : 103) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.064286 (rank : 86) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.059183 (rank : 107) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
SELPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.053575 (rank : 134) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.054421 (rank : 127) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SELV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.076941 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.074216 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.051237 (rank : 152) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.050344 (rank : 158) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SGIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.053915 (rank : 132) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.052802 (rank : 145) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.061399 (rank : 94) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
SON_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.097043 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.085853 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.087003 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.068263 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.062577 (rank : 91) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.057806 (rank : 114) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.051173 (rank : 153) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.072821 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.069682 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TAU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.053082 (rank : 142) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TB182_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.053214 (rank : 141) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.080929 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.082781 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.059280 (rank : 104) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.074936 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
TP53B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.059722 (rank : 99) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.066469 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.082582 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.084090 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H322, Q9P0H5 | Gene names | VCX2, VCX2R, VCXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 2 (VCX-B protein) (Variably charged protein X-B) (Variable charge protein on X with two repeats) (VCX-2r). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.099062 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.085242 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
VCY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.083647 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.074128 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.069601 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.050295 (rank : 159) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.057953 (rank : 113) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.065383 (rank : 78) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.067921 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.053065 (rank : 143) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZN207_HUMAN
|
||||||
| θ value | θ > 10 (rank : 197) | NC score | 0.064503 (rank : 85) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ZCHC5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N8U3 | Gene names | ZCCHC5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCHC domain-containing protein 5. | |||||
|
LDOC1_HUMAN
|
||||||
| NC score | 0.426285 (rank : 2) | θ value | 1.53129e-07 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95751 | Gene names | LDOC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LDOC1 (Leucine zipper protein down-regulated in cancer cells). | |||||
|
LDOC1_MOUSE
|
||||||
| NC score | 0.390549 (rank : 3) | θ value | 1.43324e-05 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPY9 | Gene names | Ldoc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LDOC1. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.159945 (rank : 4) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.153076 (rank : 5) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.133362 (rank : 6) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.127588 (rank : 7) | θ value | 0.00390308 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.126507 (rank : 8) | θ value | 1.87187e-05 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.124350 (rank : 9) | θ value | 0.000461057 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.119720 (rank : 10) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.118792 (rank : 11) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.116540 (rank : 12) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.116179 (rank : 13) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.111851 (rank : 14) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.111401 (rank : 15) | θ value | 0.000158464 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.110457 (rank : 16) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.109624 (rank : 17) | θ value | 0.00869519 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.109418 (rank : 18) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
NRBF2_MOUSE
|
||||||
| NC score | 0.109120 (rank : 19) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCQ3, Q9DCG3 | Gene names | Nrbf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.105449 (rank : 20) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.104322 (rank : 21) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.103613 (rank : 22) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.103413 (rank : 23) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.100461 (rank : 24) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.099062 (rank : 25) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.097324 (rank : 26) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.097043 (rank : 27) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.095910 (rank : 28) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.091464 (rank : 29) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
NRBF2_HUMAN
|
||||||
| NC score | 0.090924 (rank : 30) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96F24, Q86UR2, Q96NP6, Q9H0S9, Q9H2I2 | Gene names | NRBF2, COPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2) (Comodulator of PPAR and RXR). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.089759 (rank : 31) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.087226 (rank : 32) | θ value | 4.1701e-05 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.087003 (rank : 33) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.086592 (rank : 34) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.086458 (rank : 35) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.085853 (rank : 36) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.085242 (rank : 37) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.085220 (rank : 38) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
VCX2_HUMAN
|
||||||
| NC score | 0.084090 (rank : 39) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H322, Q9P0H5 | Gene names | VCX2, VCX2R, VCXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 2 (VCX-B protein) (Variably charged protein X-B) (Variable charge protein on X with two repeats) (VCX-2r). | |||||
|
VCY1_HUMAN
|
||||||
| NC score | 0.083647 (rank : 40) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.082781 (rank : 41) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.082582 (rank : 42) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.081440 (rank : 43) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.080929 (rank : 44) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.079089 (rank : 45) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.078470 (rank : 46) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.078375 (rank : 47) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.078219 (rank : 48) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.077913 (rank : 49) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.077872 (rank : 50) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.077448 (rank : 51) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.076941 (rank : 52) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.076384 (rank : 53) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.076097 (rank : 54) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.075130 (rank : 55) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.074936 (rank : 56) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.074495 (rank : 57) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.074216 (rank : 58) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.074128 (rank : 59) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.073857 (rank : 60) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.072821 (rank : 61) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.072159 (rank : 62) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.070765 (rank : 63) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.070442 (rank : 64) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.069924 (rank : 65) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.069682 (rank : 66) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.069601 (rank : 67) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.068786 (rank : 68) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.068626 (rank : 69) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.068553 (rank : 70) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.068263 (rank : 71) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.068162 (rank : 72) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.068083 (rank : 73) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.067921 (rank : 74) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.066749 (rank : 75) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.066469 (rank : 76) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.065773 (rank : 77) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.065383 (rank : 78) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.065351 (rank : 79) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.065158 (rank : 80) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.065025 (rank : 81) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.064861 (rank : 82) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.064826 (rank : 83) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.064823 (rank : 84) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.064503 (rank : 85) | θ value | θ > 10 (rank : 197) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.064286 (rank : 86) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
TRM6_HUMAN
|
||||||
| NC score | 0.064086 (rank : 87) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJA5, Q76P92, Q9BQV5, Q9ULR7 | Gene names | TRM6, KIAA1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.063933 (rank : 88) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.063098 (rank : 89) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.062600 (rank : 90) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.062577 (rank : 91) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.062473 (rank : 92) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.062253 (rank : 93) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.061399 (rank : 94) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.061270 (rank : 95) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.061254 (rank : 96) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.060394 (rank : 97) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.060217 (rank : 98) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.059722 (rank : 99) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.059626 (rank : 100) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.059596 (rank : 101) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.059525 (rank : 102) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.059372 (rank : 103) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.059280 (rank : 104) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
PS1C2_MOUSE
|
||||||
| NC score | 0.059265 (rank : 105) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.059241 (rank : 106) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.059183 (rank : 107) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.059031 (rank : 108) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.058614 (rank : 109) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
CEND_HUMAN
|
||||||
| NC score | 0.058612 (rank : 110) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N111, Q9NYM6 | Gene names | CEND1, BM88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle exit and neuronal differentiation protein 1 (BM88 antigen). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.058303 (rank : 111) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.058237 (rank : 112) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.057953 (rank : 113) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.057806 (rank : 114) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.057565 (rank : 115) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.057560 (rank : 116) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BAG3_HUMAN
|
||||||
| NC score | 0.057558 (rank : 117) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.057481 (rank : 118) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.057167 (rank : 119) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.057141 (rank : 120) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.056802 (rank : 121) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.055601 (rank : 122) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
CEND_MOUSE
|
||||||
| NC score | 0.054696 (rank : 123) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKC6, Q9DBA0 | Gene names | Cend1, Bm88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle exit and neuronal differentiation protein 1 (BM88 antigen). | |||||
|
PRAX_MOUSE
|
||||||
| NC score | 0.054660 (rank : 124) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
REC8L_MOUSE
|
||||||
| NC score | 0.054574 (rank : 125) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5S7, Q3UIN9, Q9JK52 | Gene names | Rec8L1, Rec8 | |||
|
Domain Architecture |

|
|||||
| Description | Meiotic recombination protein REC8-like 1 (Cohesin Rec8p). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.054453 (rank : 126) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.054421 (rank : 127) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.054319 (rank : 128) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.054061 (rank : 129) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.053980 (rank : 130) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.053964 (rank : 131) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
SGIP1_HUMAN
|
||||||
| NC score | 0.053915 (rank : 132) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.053603 (rank : 133) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
SELPL_HUMAN
|
||||||
| NC score | 0.053575 (rank : 134) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.053526 (rank : 135) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.053496 (rank : 136) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.053488 (rank : 137) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.053455 (rank : 138) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
NELFA_HUMAN
|
||||||
| NC score | 0.053370 (rank : 139) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.053223 (rank : 140) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.053214 (rank : 141) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.053082 (rank : 142) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.053065 (rank : 143) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.052932 (rank : 144) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.052802 (rank : 145) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
PHAR2_HUMAN
|
||||||
| NC score | 0.052533 (rank : 146) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.051578 (rank : 147) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.051571 (rank : 148) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.051483 (rank : 149) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.051376 (rank : 150) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.051265 (rank : 151) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.051237 (rank : 152) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.051173 (rank : 153) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.051171 (rank : 154) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
GNAS3_HUMAN
|
||||||
| NC score | 0.051073 (rank : 155) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.050467 (rank : 156) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.050358 (rank : 157) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.050344 (rank : 158) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.050295 (rank : 159) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.045230 (rank : 160) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.042952 (rank : 161) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
PREP_MOUSE
|
||||||
| NC score | 0.042523 (rank : 162) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K411, Q3THC4, Q3TJI1, Q3TPP0, Q3URQ8, Q4KUG2, Q4KUG3, Q6ZPX6, Q922N1, Q9CV63 | Gene names | Pitrm1, Kiaa1104, Ntup1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (Pitrilysin metalloproteinase 1). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.037124 (rank : 163) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.036686 (rank : 164) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
SNX21_HUMAN
|
||||||
| NC score | 0.034910 (rank : 165) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969T3, Q5JZH6, Q9BR16 | Gene names | SNX21, C20orf161, SNXL | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-21 (Sorting nexin L) (SNX-L). | |||||
|
MA1B1_HUMAN
|
||||||
| NC score | 0.034796 (rank : 166) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKM7, Q5VSG3, Q9BRS9, Q9Y5K7 | Gene names | MAN1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (EC 3.2.1.113) (ER alpha-1,2-mannosidase) (Mannosidase alpha class 1B member 1) (Man9GlcNAc2-specific-processing alpha-mannosidase). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.034155 (rank : 167) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
IRF7_MOUSE
|
||||||
| NC score | 0.034093 (rank : 168) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70434 | Gene names | Irf7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.033988 (rank : 169) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
ES8L2_HUMAN
|
||||||
| NC score | 0.033928 (rank : 170) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H6S3, Q53GM8, Q8WYW7, Q96K06, Q9H6K9 | Gene names | EPS8L2, EPS8R2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
FOSB_MOUSE
|
||||||
| NC score | 0.033632 (rank : 171) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
FOSL2_HUMAN
|
||||||
| NC score | 0.033518 (rank : 172) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
ZCHC6_HUMAN
|
||||||
| NC score | 0.031278 (rank : 173) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZCHC6_MOUSE
|
||||||
| NC score | 0.029771 (rank : 174) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
CF057_HUMAN
|
||||||
| NC score | 0.029738 (rank : 175) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VUM1 | Gene names | C6orf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0369 protein C6orf57 precursor. | |||||
|
CU030_HUMAN
|
||||||
| NC score | 0.029486 (rank : 176) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UFM2 | Gene names | C21orf30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C21orf30. | |||||
|
HSF1_HUMAN
|
||||||
| NC score | 0.028875 (rank : 177) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.028793 (rank : 178) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
CNBP_MOUSE
|
||||||
| NC score | 0.028116 (rank : 179) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
LTB1L_HUMAN
|
||||||
| NC score | 0.027174 (rank : 180) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
SHD_HUMAN
|
||||||
| NC score | 0.026953 (rank : 181) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96IW2, Q96NC2 | Gene names | SHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein D. | |||||
|
LTB1S_HUMAN
|
||||||
| NC score | 0.025511 (rank : 182) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.024999 (rank : 183) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
IZUM1_MOUSE
|
||||||
| NC score | 0.022866 (rank : 184) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9J7 | Gene names | Izumo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Izumo sperm-egg fusion protein 1 precursor (Sperm-specific protein izumo). | |||||
|
MCM3A_HUMAN
|
||||||
| NC score | 0.022069 (rank : 185) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.020712 (rank : 186) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
ATS12_HUMAN
|
||||||
| NC score | 0.019498 (rank : 187) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
GAK18_HUMAN
|
||||||
| NC score | 0.018015 (rank : 188) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.017868 (rank : 189) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.017668 (rank : 190) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.017538 (rank : 191) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
PO2F2_MOUSE
|
||||||
| NC score | 0.014970 (rank : 192) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00196, Q00197, Q00198, Q00199, Q00200, Q00201, Q05882, Q61995, Q61996, Q64245 | Gene names | Pou2f2, Oct2, Otf2 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.014477 (rank : 193) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
ZDHC9_HUMAN
|
||||||
| NC score | 0.013050 (rank : 194) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y397, Q59EK4, Q5JSW5, Q8WWS7, Q9BPY4, Q9NSP0, Q9NVL0, Q9NVR6 | Gene names | ZDHHC9, ZNF379 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9) (Zinc finger protein 379). | |||||
|
ITA5_MOUSE
|
||||||
| NC score | 0.009302 (rank : 195) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11688 | Gene names | Itga5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
BAI2_MOUSE
|
||||||
| NC score | 0.009235 (rank : 196) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGM1, Q3TYC8, Q3UN11, Q3UNE2, Q6PGN0 | Gene names | Bai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
SRMS_MOUSE
|
||||||
| NC score | 0.000750 (rank : 197) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62270, Q62360, Q923M5 | Gene names | Srms, Srm | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2) (PTK70). | |||||