Please be patient as the page loads
|
NACA_MOUSE
|
||||||
| SwissProt Accessions | Q60817 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (Alpha-NAC) (Alpha-NAC/1.9.2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NACA_HUMAN
|
||||||
| θ value | 3.56116e-89 (rank : 1) | NC score | 0.999334 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13765, Q53A18, Q53G46 | Gene names | NACA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (NAC-alpha) (Alpha-NAC) (Hom s 2.02). | |||||
|
NACA_MOUSE
|
||||||
| θ value | 3.56116e-89 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60817 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (Alpha-NAC) (Alpha-NAC/1.9.2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 9.0981e-77 (rank : 3) | NC score | 0.374885 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BTF3_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 4) | NC score | 0.184155 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20290, Q13893, Q76M56 | Gene names | BTF3, NACB | |||
|
Domain Architecture |
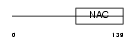
|
|||||
| Description | Transcription factor BTF3 (RNA polymerase B transcription factor 3). | |||||
|
BTF3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 5) | NC score | 0.179888 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64152, Q64153, Q6GU63, Q6P3F3 | Gene names | Btf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor BTF3 (RNA polymerase B transcription factor 3). | |||||
|
SDPR_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.116405 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.110363 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
BT3L1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.164338 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13890 | Gene names | BTF3L1 | |||
|
Domain Architecture |
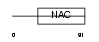
|
|||||
| Description | Transcription factor BTF3 homolog 1 (Basic transcription factor 3-like 1). | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.039460 (rank : 76) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
DEK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.066932 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
PO5F1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.014913 (rank : 81) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01860, P31359, Q15167, Q15168, Q16422 | Gene names | POU5F1, OCT3, OCT4, OTF3 | |||
|
Domain Architecture |
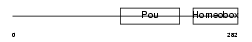
|
|||||
| Description | POU domain, class 5, transcription factor 1 (Octamer-binding transcription factor 3) (Oct-3) (Oct-4). | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.037920 (rank : 77) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.014732 (rank : 82) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
FAN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.019060 (rank : 80) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35242 | Gene names | Nsmaf, Fan | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAN (Factor associated with N-SMase activation) (Factor associated with neutral sphingomyelinase activation). | |||||
|
ZN131_MOUSE
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.005520 (rank : 87) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3J5, Q3UYZ4, Q80UU7, Q8C8D0, Q9D042 | Gene names | Znf131, Zfp131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 131. | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.032542 (rank : 78) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.002673 (rank : 90) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
HXK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.014301 (rank : 83) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17710, Q61659, Q64476, Q64479 | Gene names | Hk1 | |||
|
Domain Architecture |

|
|||||
| Description | Hexokinase-1 (EC 2.7.1.1) (Hexokinase type I) (HK I) (Hexokinase, tumor isozyme). | |||||
|
K2C1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.007276 (rank : 85) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.005158 (rank : 88) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
UN119_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.032010 (rank : 79) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13432, O95126 | Gene names | UNC119, RG4 | |||
|
Domain Architecture |
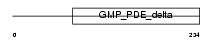
|
|||||
| Description | Unc-119 protein homolog (Retinal protein 4) (HRG4). | |||||
|
ZN131_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.005013 (rank : 89) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52739, Q6PIF0 | Gene names | ZNF131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 131. | |||||
|
GALT5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.013974 (rank : 84) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.006775 (rank : 86) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
ALEX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.064223 (rank : 29) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.053211 (rank : 59) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
AP180_MOUSE
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.050837 (rank : 70) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.070089 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.050520 (rank : 72) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BT3L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.075773 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13891 | Gene names | BTF3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor BTF3 homolog 2 (Basic transcription factor 3-like 2). | |||||
|
BT3L3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.076085 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13892 | Gene names | BTF3L3 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor BTF3 homolog 3 (Basic transcription factor 3-like 3). | |||||
|
BT3L4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.075937 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96K17 | Gene names | BTF3L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor BTF3 homolog 4 (Basic transcription factor 3-like 4). | |||||
|
BT3L4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.075937 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQH7 | Gene names | Btf3l4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor BTF3 homolog 4 (Basic transcription factor 3-like 4). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.055977 (rank : 54) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CJ095_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.052257 (rank : 62) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9H7T3 | Gene names | C10orf95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf95. | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.050149 (rank : 75) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.060838 (rank : 37) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.059294 (rank : 42) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.061278 (rank : 36) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.068313 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
H14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.050858 (rank : 69) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
K0460_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.052786 (rank : 60) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
K1683_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.056222 (rank : 52) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
KI67_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.061710 (rank : 32) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.061367 (rank : 34) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.075632 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.064740 (rank : 28) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.077439 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.067850 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.052360 (rank : 61) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.050900 (rank : 68) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.053217 (rank : 58) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.067822 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.056174 (rank : 53) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.082396 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MUCL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.060121 (rank : 39) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96DR8, Q32ZB5 | Gene names | SBEM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small breast epithelial mucin precursor (Protein BS106). | |||||
|
NELFA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.053299 (rank : 56) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.050665 (rank : 71) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.067078 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NU214_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.059160 (rank : 44) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
NUP62_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.056231 (rank : 51) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
PARM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.061347 (rank : 35) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PRAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.062136 (rank : 31) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRAX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.053291 (rank : 57) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRCC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.060758 (rank : 38) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.090785 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.074503 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.051953 (rank : 66) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.058021 (rank : 46) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.057737 (rank : 48) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.059206 (rank : 43) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.050231 (rank : 73) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050181 (rank : 74) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
SDC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.056389 (rank : 50) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75056, Q5T1Z6, Q5T1Z7, Q96CT3, Q96PR8 | Gene names | SDC3, KIAA0468 | |||
|
Domain Architecture |
|
|||||
| Description | Syndecan-3 (SYND3). | |||||
|
SDC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.051929 (rank : 67) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q64519, Q3UDD9, Q6ZQA4, Q7TQD4 | Gene names | Sdc3, Kiaa0468 | |||
|
Domain Architecture |
|
|||||
| Description | Syndecan-3 precursor (SYND3). | |||||
|
SELV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.073975 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
SNPC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.052178 (rank : 63) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q13487, Q13486 | Gene names | SNAPC2, SNAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2) (Proximal sequence element-binding transcription factor subunit delta) (PSE- binding factor subunit delta) (PTF subunit delta). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.056720 (rank : 49) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.058624 (rank : 45) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.061528 (rank : 33) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.059401 (rank : 40) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.057882 (rank : 47) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.066887 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.078412 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TM108_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.059400 (rank : 41) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.053335 (rank : 55) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
UBN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.052006 (rank : 64) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
VCY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.062899 (rank : 30) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.051993 (rank : 65) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.065288 (rank : 27) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
NACA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.56116e-89 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60817 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (Alpha-NAC) (Alpha-NAC/1.9.2). | |||||
|
NACA_HUMAN
|
||||||
| NC score | 0.999334 (rank : 2) | θ value | 3.56116e-89 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13765, Q53A18, Q53G46 | Gene names | NACA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (NAC-alpha) (Alpha-NAC) (Hom s 2.02). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.374885 (rank : 3) | θ value | 9.0981e-77 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BTF3_HUMAN
|
||||||
| NC score | 0.184155 (rank : 4) | θ value | 0.0113563 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20290, Q13893, Q76M56 | Gene names | BTF3, NACB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor BTF3 (RNA polymerase B transcription factor 3). | |||||
|
BTF3_MOUSE
|
||||||
| NC score | 0.179888 (rank : 5) | θ value | 0.0330416 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64152, Q64153, Q6GU63, Q6P3F3 | Gene names | Btf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor BTF3 (RNA polymerase B transcription factor 3). | |||||
|
BT3L1_HUMAN
|
||||||
| NC score | 0.164338 (rank : 6) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13890 | Gene names | BTF3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor BTF3 homolog 1 (Basic transcription factor 3-like 1). | |||||
|
SDPR_HUMAN
|
||||||
| NC score | 0.116405 (rank : 7) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.110363 (rank : 8) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.090785 (rank : 9) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.082396 (rank : 10) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.078412 (rank : 11) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.077439 (rank : 12) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
BT3L3_HUMAN
|
||||||
| NC score | 0.076085 (rank : 13) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13892 | Gene names | BTF3L3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor BTF3 homolog 3 (Basic transcription factor 3-like 3). | |||||
|
BT3L4_HUMAN
|
||||||
| NC score | 0.075937 (rank : 14) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96K17 | Gene names | BTF3L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor BTF3 homolog 4 (Basic transcription factor 3-like 4). | |||||
|
BT3L4_MOUSE
|
||||||
| NC score | 0.075937 (rank : 15) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQH7 | Gene names | Btf3l4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor BTF3 homolog 4 (Basic transcription factor 3-like 4). | |||||
|
BT3L2_HUMAN
|
||||||
| NC score | 0.075773 (rank : 16) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13891 | Gene names | BTF3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor BTF3 homolog 2 (Basic transcription factor 3-like 2). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.075632 (rank : 17) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.074503 (rank : 18) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.073975 (rank : 19) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.070089 (rank : 20) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.068313 (rank : 21) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.067850 (rank : 22) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.067822 (rank : 23) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.067078 (rank : 24) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.066932 (rank : 25) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.066887 (rank : 26) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.065288 (rank : 27) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.064740 (rank : 28) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.064223 (rank : 29) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
VCY1_HUMAN
|
||||||
| NC score | 0.062899 (rank : 30) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.062136 (rank : 31) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.061710 (rank : 32) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.061528 (rank : 33) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.061367 (rank : 34) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.061347 (rank : 35) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.061278 (rank : 36) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.060838 (rank : 37) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
PRCC_HUMAN
|
||||||
| NC score | 0.060758 (rank : 38) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
MUCL_HUMAN
|
||||||
| NC score | 0.060121 (rank : 39) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96DR8, Q32ZB5 | Gene names | SBEM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small breast epithelial mucin precursor (Protein BS106). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.059401 (rank : 40) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.059400 (rank : 41) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.059294 (rank : 42) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.059206 (rank : 43) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.059160 (rank : 44) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.058624 (rank : 45) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.058021 (rank : 46) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.057882 (rank : 47) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.057737 (rank : 48) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.056720 (rank : 49) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SDC3_HUMAN
|
||||||
| NC score | 0.056389 (rank : 50) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75056, Q5T1Z6, Q5T1Z7, Q96CT3, Q96PR8 | Gene names | SDC3, KIAA0468 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-3 (SYND3). | |||||
|
NUP62_HUMAN
|
||||||
| NC score | 0.056231 (rank : 51) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.056222 (rank : 52) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.056174 (rank : 53) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.055977 (rank : 54) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.053335 (rank : 55) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
NELFA_HUMAN
|
||||||
| NC score | 0.053299 (rank : 56) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
PRAX_MOUSE
|
||||||
| NC score | 0.053291 (rank : 57) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.053217 (rank : 58) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.053211 (rank : 59) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.052786 (rank : 60) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.052360 (rank : 61) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CJ095_HUMAN
|
||||||
| NC score | 0.052257 (rank : 62) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9H7T3 | Gene names | C10orf95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf95. | |||||
|
SNPC2_HUMAN
|
||||||
| NC score | 0.052178 (rank : 63) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q13487, Q13486 | Gene names | SNAPC2, SNAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2) (Proximal sequence element-binding transcription factor subunit delta) (PSE- binding factor subunit delta) (PTF subunit delta). | |||||
|
UBN1_HUMAN
|
||||||
| NC score | 0.052006 (rank : 64) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.051993 (rank : 65) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.051953 (rank : 66) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SDC3_MOUSE
|
||||||
| NC score | 0.051929 (rank : 67) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q64519, Q3UDD9, Q6ZQA4, Q7TQD4 | Gene names | Sdc3, Kiaa0468 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-3 precursor (SYND3). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.050900 (rank : 68) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
H14_HUMAN
|
||||||
| NC score | 0.050858 (rank : 69) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
AP180_MOUSE
|
||||||
| NC score | 0.050837 (rank : 70) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.050665 (rank : 71) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.050520 (rank : 72) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.050231 (rank : 73) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.050181 (rank : 74) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.050149 (rank : 75) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.039460 (rank : 76) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.037920 (rank : 77) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.032542 (rank : 78) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
UN119_HUMAN
|
||||||
| NC score | 0.032010 (rank : 79) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13432, O95126 | Gene names | UNC119, RG4 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-119 protein homolog (Retinal protein 4) (HRG4). | |||||
|
FAN_MOUSE
|
||||||
| NC score | 0.019060 (rank : 80) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35242 | Gene names | Nsmaf, Fan | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAN (Factor associated with N-SMase activation) (Factor associated with neutral sphingomyelinase activation). | |||||
|
PO5F1_HUMAN
|
||||||
| NC score | 0.014913 (rank : 81) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01860, P31359, Q15167, Q15168, Q16422 | Gene names | POU5F1, OCT3, OCT4, OTF3 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 5, transcription factor 1 (Octamer-binding transcription factor 3) (Oct-3) (Oct-4). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.014732 (rank : 82) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
HXK1_MOUSE
|
||||||
| NC score | 0.014301 (rank : 83) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17710, Q61659, Q64476, Q64479 | Gene names | Hk1 | |||
|
Domain Architecture |

|
|||||
| Description | Hexokinase-1 (EC 2.7.1.1) (Hexokinase type I) (HK I) (Hexokinase, tumor isozyme). | |||||
|
GALT5_HUMAN
|
||||||
| NC score | 0.013974 (rank : 84) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
K2C1_HUMAN
|
||||||
| NC score | 0.007276 (rank : 85) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.006775 (rank : 86) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
ZN131_MOUSE
|
||||||
| NC score | 0.005520 (rank : 87) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3J5, Q3UYZ4, Q80UU7, Q8C8D0, Q9D042 | Gene names | Znf131, Zfp131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 131. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.005158 (rank : 88) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ZN131_HUMAN
|
||||||
| NC score | 0.005013 (rank : 89) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52739, Q6PIF0 | Gene names | ZNF131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 131. | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.002673 (rank : 90) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||