Please be patient as the page loads
|
ATAD2_MOUSE
|
||||||
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ATAD2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.979627 (rank : 2) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 167 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
TERA_MOUSE
|
||||||
| θ value | 1.7238e-59 (rank : 3) | NC score | 0.902401 (rank : 3) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_HUMAN
|
||||||
| θ value | 2.25136e-59 (rank : 4) | NC score | 0.902362 (rank : 4) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PRS4_HUMAN
|
||||||
| θ value | 1.28137e-46 (rank : 5) | NC score | 0.897974 (rank : 7) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| θ value | 1.28137e-46 (rank : 6) | NC score | 0.897974 (rank : 8) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS7_HUMAN
|
||||||
| θ value | 2.04573e-44 (rank : 7) | NC score | 0.897450 (rank : 9) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| θ value | 2.04573e-44 (rank : 8) | NC score | 0.897440 (rank : 10) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 1.01529e-43 (rank : 9) | NC score | 0.887425 (rank : 19) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS10_HUMAN
|
||||||
| θ value | 1.32601e-43 (rank : 10) | NC score | 0.898332 (rank : 5) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| θ value | 1.32601e-43 (rank : 11) | NC score | 0.898332 (rank : 6) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS8_HUMAN
|
||||||
| θ value | 1.73182e-43 (rank : 12) | NC score | 0.896276 (rank : 11) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| θ value | 1.73182e-43 (rank : 13) | NC score | 0.896276 (rank : 12) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 6.58091e-43 (rank : 14) | NC score | 0.886130 (rank : 21) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS6A_HUMAN
|
||||||
| θ value | 1.05066e-40 (rank : 15) | NC score | 0.894111 (rank : 14) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6A_MOUSE
|
||||||
| θ value | 1.3722e-40 (rank : 16) | NC score | 0.894334 (rank : 13) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
KATL1_HUMAN
|
||||||
| θ value | 3.73686e-38 (rank : 17) | NC score | 0.877293 (rank : 22) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_MOUSE
|
||||||
| θ value | 4.568e-36 (rank : 18) | NC score | 0.876156 (rank : 23) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 7.79185e-36 (rank : 19) | NC score | 0.886312 (rank : 20) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
ATAD1_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 20) | NC score | 0.887726 (rank : 17) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 21) | NC score | 0.887726 (rank : 18) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
PRS6B_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 22) | NC score | 0.888328 (rank : 15) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| θ value | 2.50655e-34 (rank : 23) | NC score | 0.887886 (rank : 16) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 2.50655e-34 (rank : 24) | NC score | 0.864379 (rank : 36) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 2.50655e-34 (rank : 25) | NC score | 0.864363 (rank : 37) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 9.52487e-34 (rank : 26) | NC score | 0.874365 (rank : 24) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 2.12192e-33 (rank : 27) | NC score | 0.873893 (rank : 25) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 3.61944e-33 (rank : 28) | NC score | 0.865930 (rank : 32) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 3.61944e-33 (rank : 29) | NC score | 0.864688 (rank : 35) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
AFG31_MOUSE
|
||||||
| θ value | 6.17384e-33 (rank : 30) | NC score | 0.867670 (rank : 30) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
AFG32_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 31) | NC score | 0.863836 (rank : 38) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
KTNA1_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 32) | NC score | 0.871140 (rank : 26) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
AFG32_MOUSE
|
||||||
| θ value | 1.0531e-32 (rank : 33) | NC score | 0.863426 (rank : 39) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
NSF_MOUSE
|
||||||
| θ value | 6.82597e-32 (rank : 34) | NC score | 0.865517 (rank : 33) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
SPAST_HUMAN
|
||||||
| θ value | 1.16434e-31 (rank : 35) | NC score | 0.868094 (rank : 29) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 36) | NC score | 0.866736 (rank : 31) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
KTNA1_MOUSE
|
||||||
| θ value | 1.98606e-31 (rank : 37) | NC score | 0.868947 (rank : 28) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
NSF_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 38) | NC score | 0.864963 (rank : 34) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
SPG7_HUMAN
|
||||||
| θ value | 9.8567e-31 (rank : 39) | NC score | 0.869589 (rank : 27) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | 1.4233e-29 (rank : 40) | NC score | 0.863233 (rank : 40) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
YMEL1_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 41) | NC score | 0.859634 (rank : 41) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 1.99067e-23 (rank : 42) | NC score | 0.826511 (rank : 42) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 9.87957e-23 (rank : 43) | NC score | 0.821457 (rank : 43) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 44) | NC score | 0.221181 (rank : 76) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 45) | NC score | 0.204835 (rank : 85) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
BRD1_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 46) | NC score | 0.214532 (rank : 79) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 47) | NC score | 0.148937 (rank : 93) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 48) | NC score | 0.214734 (rank : 78) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 49) | NC score | 0.216464 (rank : 77) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 50) | NC score | 0.234881 (rank : 73) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 51) | NC score | 0.233225 (rank : 74) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
BRWD1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 52) | NC score | 0.148756 (rank : 95) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
BCS1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 53) | NC score | 0.639969 (rank : 46) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 54) | NC score | 0.257821 (rank : 61) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 55) | NC score | 0.246847 (rank : 66) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
TAF1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 56) | NC score | 0.257247 (rank : 63) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
BCS1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 57) | NC score | 0.630438 (rank : 47) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
BRD7_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 58) | NC score | 0.268944 (rank : 56) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
BRD7_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 59) | NC score | 0.264503 (rank : 57) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 60) | NC score | 0.565695 (rank : 50) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 61) | NC score | 0.573280 (rank : 49) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 62) | NC score | 0.213822 (rank : 80) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BRD3_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 63) | NC score | 0.257585 (rank : 62) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 64) | NC score | 0.195031 (rank : 86) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
TAF1L_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 65) | NC score | 0.246232 (rank : 69) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 66) | NC score | 0.205555 (rank : 83) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 67) | NC score | 0.247133 (rank : 65) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 68) | NC score | 0.194682 (rank : 87) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 69) | NC score | 0.205378 (rank : 84) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
GCNL2_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 70) | NC score | 0.246648 (rank : 67) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
ATD3A_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 71) | NC score | 0.580844 (rank : 48) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
PB1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 72) | NC score | 0.237548 (rank : 72) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 73) | NC score | 0.102999 (rank : 101) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 74) | NC score | 0.241150 (rank : 71) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 75) | NC score | 0.101129 (rank : 105) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
TRP13_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 76) | NC score | 0.653432 (rank : 45) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
TRP13_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 77) | NC score | 0.655707 (rank : 44) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 78) | NC score | 0.258041 (rank : 60) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
BRDT_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 79) | NC score | 0.244306 (rank : 70) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
BRD9_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 80) | NC score | 0.259334 (rank : 59) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
BRD9_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 81) | NC score | 0.260294 (rank : 58) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
BRDT_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 82) | NC score | 0.246488 (rank : 68) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
PCAF_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 83) | NC score | 0.253812 (rank : 64) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92831, Q6NSK1 | Gene names | PCAF | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase PCAF (EC 2.3.1.48) (P300/CBP-associated factor) (P/CAF) (Histone acetylase PCAF). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 84) | NC score | 0.222117 (rank : 75) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 85) | NC score | 0.172228 (rank : 92) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 86) | NC score | 0.174839 (rank : 89) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 87) | NC score | 0.146307 (rank : 96) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 88) | NC score | 0.143770 (rank : 97) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 89) | NC score | 0.183929 (rank : 88) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 90) | NC score | 0.040785 (rank : 126) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 91) | NC score | 0.172715 (rank : 91) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 92) | NC score | 0.016629 (rank : 147) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 93) | NC score | 0.174440 (rank : 90) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 94) | NC score | 0.394978 (rank : 51) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 95) | NC score | 0.392676 (rank : 52) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 96) | NC score | 0.043106 (rank : 124) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 97) | NC score | 0.148845 (rank : 94) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
F10A5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 98) | NC score | 0.020283 (rank : 142) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
MNDA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 99) | NC score | 0.017500 (rank : 145) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |
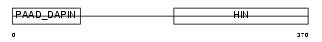
|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
CDR2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 100) | NC score | 0.027599 (rank : 133) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97817 | Gene names | Cdr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2. | |||||
|
LONM_HUMAN
|
||||||
| θ value | 0.47712 (rank : 101) | NC score | 0.386378 (rank : 53) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
POL2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 102) | NC score | 0.028862 (rank : 132) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11369, Q60713, Q61787 | Gene names | Pol | |||
|
Domain Architecture |

|
|||||
| Description | Retrovirus-related Pol polyprotein LINE-1 (Long interspersed element- 1) (L1) [Includes: Reverse transcriptase (EC 2.7.7.49); Endonuclease]. | |||||
|
RUVB1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 103) | NC score | 0.285614 (rank : 54) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 104) | NC score | 0.285578 (rank : 55) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 0.47712 (rank : 105) | NC score | 0.141613 (rank : 98) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 0.47712 (rank : 106) | NC score | 0.139563 (rank : 99) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 107) | NC score | 0.011788 (rank : 156) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
PPHLN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 108) | NC score | 0.023564 (rank : 138) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2H1, Q8BUZ6 | Gene names | Pphln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1. | |||||
|
KIF3C_HUMAN
|
||||||
| θ value | 0.813845 (rank : 109) | NC score | 0.007898 (rank : 173) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |
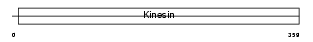
|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
KIF3C_MOUSE
|
||||||
| θ value | 0.813845 (rank : 110) | NC score | 0.007938 (rank : 172) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35066, O35229 | Gene names | Kif3c | |||
|
Domain Architecture |
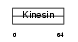
|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
SYEP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 111) | NC score | 0.015956 (rank : 149) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 1.06291 (rank : 112) | NC score | 0.006859 (rank : 174) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MRP6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 113) | NC score | 0.009632 (rank : 164) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95255, P78420, Q9UMZ7 | Gene names | ABCC6, ARA, MRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance-associated protein 6 (ATP-binding cassette sub- family C member 6) (Anthracycline resistance-associated protein) (Multi-specific organic anion transporter-E) (MOAT-E). | |||||
|
NAT10_HUMAN
|
||||||
| θ value | 1.06291 (rank : 114) | NC score | 0.022432 (rank : 139) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0A0, Q86WK5, Q9C0F4, Q9HA61, Q9NVF2 | Gene names | NAT10, ALP, KIAA1709 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 10 (EC 2.3.1.-). | |||||
|
NCPR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 115) | NC score | 0.018952 (rank : 144) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16435, Q16455, Q8N181, Q9H3M8 | Gene names | POR, CYPOR | |||
|
Domain Architecture |

|
|||||
| Description | NADPH--cytochrome P450 reductase (EC 1.6.2.4) (CPR) (P450R). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 116) | NC score | 0.042634 (rank : 125) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 117) | NC score | 0.012252 (rank : 155) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CF165_HUMAN
|
||||||
| θ value | 1.81305 (rank : 118) | NC score | 0.020584 (rank : 141) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYR0, Q8N9U4 | Gene names | C6orf165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf165. | |||||
|
DNJC4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 119) | NC score | 0.009089 (rank : 167) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |
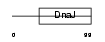
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
JPH4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 120) | NC score | 0.010549 (rank : 159) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
K1H6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 121) | NC score | 0.004141 (rank : 180) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O76013 | Gene names | KRTHA6, HHA6, HKA6 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha6 (Hair keratin, type I Ha6). | |||||
|
RFC4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 122) | NC score | 0.101617 (rank : 103) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RFC4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 123) | NC score | 0.091670 (rank : 107) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
SETB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 124) | NC score | 0.016729 (rank : 146) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 125) | NC score | 0.019745 (rank : 143) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 126) | NC score | 0.030172 (rank : 131) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LIN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 127) | NC score | 0.026465 (rank : 135) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08547 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LINE-1 reverse transcriptase homolog. | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 128) | NC score | 0.211964 (rank : 81) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 129) | NC score | 0.031346 (rank : 130) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 130) | NC score | 0.009243 (rank : 166) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
LCP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 131) | NC score | 0.010324 (rank : 161) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60787 | Gene names | Lcp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 132) | NC score | 0.034000 (rank : 127) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 133) | NC score | 0.008291 (rank : 171) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 134) | NC score | 0.012261 (rank : 153) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
STAR7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 135) | NC score | 0.014033 (rank : 151) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 136) | NC score | 0.010707 (rank : 158) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CDR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 137) | NC score | 0.020806 (rank : 140) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01850, Q13977 | Gene names | CDR2, PCD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2 (Paraneoplastic cerebellar degeneration-associated antigen) (Major Yo paraneoplastic antigen). | |||||
|
COQ9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 138) | NC score | 0.015101 (rank : 150) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75208, Q7L5V7, Q7Z5T6, Q8NBL4, Q9NTJ2, Q9P056 | Gene names | COQ9, C16orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquinone biosynthesis protein COQ9, mitochondrial precursor. | |||||
|
CT032_HUMAN
|
||||||
| θ value | 4.03905 (rank : 139) | NC score | 0.010136 (rank : 162) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NQ75, Q96K09, Q9BYL5 | Gene names | HEFL, C20orf32 | |||
|
Domain Architecture |
|
|||||
| Description | HEF-like protein. | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 140) | NC score | 0.002668 (rank : 181) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 141) | NC score | 0.023652 (rank : 137) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
REST_MOUSE
|
||||||
| θ value | 4.03905 (rank : 142) | NC score | 0.002460 (rank : 182) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
TF3C4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 143) | NC score | 0.009976 (rank : 163) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BMQ2, Q8BKZ4 | Gene names | Gtf3c4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 4 (EC 2.3.1.48) (Transcription factor IIIC subunit delta) (TF3C-delta) (TFIIIC 90 kDa subunit) (TFIIIC 90). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 144) | NC score | 0.026834 (rank : 134) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ZN518_HUMAN
|
||||||
| θ value | 4.03905 (rank : 145) | NC score | -0.003973 (rank : 187) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6AHZ1, O15044, Q32MP4 | Gene names | ZNF518, KIAA0335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 518. | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 146) | NC score | 0.032645 (rank : 128) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
JPH4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 147) | NC score | 0.008551 (rank : 170) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 148) | NC score | 0.008555 (rank : 169) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 149) | NC score | 0.005601 (rank : 178) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 150) | NC score | 0.000429 (rank : 184) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 5.27518 (rank : 151) | NC score | 0.025665 (rank : 136) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
RB15B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 152) | NC score | 0.004388 (rank : 179) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
SYBU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 153) | NC score | 0.006539 (rank : 176) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHS8, Q3TQN7, Q401F3, Q401F4, Q571C1, Q6P1J2, Q80XH0, Q8BHS7, Q8BI27 | Gene names | Sybu, Golsyn, Kiaa1472 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntabulin (Syntaxin-1-binding protein) (Golgi-localized syntaphilin- related protein) (m-Golsyn). | |||||
|
CF010_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.010940 (rank : 157) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | -0.000314 (rank : 186) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.031700 (rank : 129) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
K1C16_MOUSE
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.001802 (rank : 183) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2K1 | Gene names | Krt16, Krt1-16 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
KIF14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.008650 (rank : 168) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
NCOA7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.006416 (rank : 177) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6DFV7, Q3TAR1, Q3UNB3, Q66K05, Q6ZQM4, Q8BJ15, Q8BJ39 | Gene names | Ncoa7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7. | |||||
|
STK10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.000092 (rank : 185) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.009505 (rank : 165) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
COQ9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.012258 (rank : 154) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1Z0, Q9CT61 | Gene names | Coq9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquinone biosynthesis protein COQ9, mitochondrial precursor. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.013330 (rank : 152) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.016089 (rank : 148) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
STAR7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.010543 (rank : 160) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
UT14B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.006679 (rank : 175) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
WRIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.205797 (rank : 82) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.054925 (rank : 118) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.053970 (rank : 119) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
KAD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.084779 (rank : 110) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
KAD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.065482 (rank : 112) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
LY10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.064102 (rank : 114) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.050277 (rank : 123) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.050523 (rank : 122) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
RFC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.102315 (rank : 102) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.106574 (rank : 100) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RFC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.056061 (rank : 117) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.056583 (rank : 116) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.059542 (rank : 115) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
RUVB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.101304 (rank : 104) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.098719 (rank : 106) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
THNSL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.087670 (rank : 109) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
THNSL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.090946 (rank : 108) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
TIF1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.052926 (rank : 120) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TIF1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.051889 (rank : 121) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.065115 (rank : 113) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.065805 (rank : 111) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 167 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.979627 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
TERA_MOUSE
|
||||||
| NC score | 0.902401 (rank : 3) | θ value | 1.7238e-59 (rank : 3) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_HUMAN
|
||||||
| NC score | 0.902362 (rank : 4) | θ value | 2.25136e-59 (rank : 4) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PRS10_HUMAN
|
||||||
| NC score | 0.898332 (rank : 5) | θ value | 1.32601e-43 (rank : 10) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| NC score | 0.898332 (rank : 6) | θ value | 1.32601e-43 (rank : 11) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS4_HUMAN
|
||||||
| NC score | 0.897974 (rank : 7) | θ value | 1.28137e-46 (rank : 5) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| NC score | 0.897974 (rank : 8) | θ value | 1.28137e-46 (rank : 6) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS7_HUMAN
|
||||||
| NC score | 0.897450 (rank : 9) | θ value | 2.04573e-44 (rank : 7) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| NC score | 0.897440 (rank : 10) | θ value | 2.04573e-44 (rank : 8) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS8_HUMAN
|
||||||
| NC score | 0.896276 (rank : 11) | θ value | 1.73182e-43 (rank : 12) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| NC score | 0.896276 (rank : 12) | θ value | 1.73182e-43 (rank : 13) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS6A_MOUSE
|
||||||
| NC score | 0.894334 (rank : 13) | θ value | 1.3722e-40 (rank : 16) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6A_HUMAN
|
||||||
| NC score | 0.894111 (rank : 14) | θ value | 1.05066e-40 (rank : 15) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6B_HUMAN
|
||||||
| NC score | 0.888328 (rank : 15) | θ value | 1.12514e-34 (rank : 22) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| NC score | 0.887886 (rank : 16) | θ value | 2.50655e-34 (rank : 23) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
ATAD1_HUMAN
|
||||||
| NC score | 0.887726 (rank : 17) | θ value | 1.12514e-34 (rank : 20) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| NC score | 0.887726 (rank : 18) | θ value | 1.12514e-34 (rank : 21) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.887425 (rank : 19) | θ value | 1.01529e-43 (rank : 9) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.886312 (rank : 20) | θ value | 7.79185e-36 (rank : 19) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.886130 (rank : 21) | θ value | 6.58091e-43 (rank : 14) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
KATL1_HUMAN
|
||||||
| NC score | 0.877293 (rank : 22) | θ value | 3.73686e-38 (rank : 17) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_MOUSE
|
||||||
| NC score | 0.876156 (rank : 23) | θ value | 4.568e-36 (rank : 18) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.874365 (rank : 24) | θ value | 9.52487e-34 (rank : 26) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.873893 (rank : 25) | θ value | 2.12192e-33 (rank : 27) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
KTNA1_HUMAN
|
||||||
| NC score | 0.871140 (rank : 26) | θ value | 8.06329e-33 (rank : 32) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
SPG7_HUMAN
|
||||||
| NC score | 0.869589 (rank : 27) | θ value | 9.8567e-31 (rank : 39) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
KTNA1_MOUSE
|
||||||
| NC score | 0.868947 (rank : 28) | θ value | 1.98606e-31 (rank : 37) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
SPAST_HUMAN
|
||||||
| NC score | 0.868094 (rank : 29) | θ value | 1.16434e-31 (rank : 35) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
AFG31_MOUSE
|
||||||
| NC score | 0.867670 (rank : 30) | θ value | 6.17384e-33 (rank : 30) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.866736 (rank : 31) | θ value | 1.16434e-31 (rank : 36) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.865930 (rank : 32) | θ value | 3.61944e-33 (rank : 28) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
NSF_MOUSE
|
||||||
| NC score | 0.865517 (rank : 33) | θ value | 6.82597e-32 (rank : 34) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
NSF_HUMAN
|
||||||
| NC score | 0.864963 (rank : 34) | θ value | 1.98606e-31 (rank : 38) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.864688 (rank : 35) | θ value | 3.61944e-33 (rank : 29) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.864379 (rank : 36) | θ value | 2.50655e-34 (rank : 24) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.864363 (rank : 37) | θ value | 2.50655e-34 (rank : 25) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
AFG32_HUMAN
|
||||||
| NC score | 0.863836 (rank : 38) | θ value | 8.06329e-33 (rank : 31) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| NC score | 0.863426 (rank : 39) | θ value | 1.0531e-32 (rank : 33) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.863233 (rank : 40) | θ value | 1.4233e-29 (rank : 40) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
YMEL1_HUMAN
|
||||||
| NC score | 0.859634 (rank : 41) | θ value | 1.33217e-27 (rank : 41) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.826511 (rank : 42) | θ value | 1.99067e-23 (rank : 42) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.821457 (rank : 43) | θ value | 9.87957e-23 (rank : 43) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
TRP13_MOUSE
|
||||||
| NC score | 0.655707 (rank : 44) | θ value | 2.44474e-05 (rank : 77) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| NC score | 0.653432 (rank : 45) | θ value | 2.44474e-05 (rank : 76) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
BCS1_HUMAN
|
||||||
| NC score | 0.639969 (rank : 46) | θ value | 7.59969e-07 (rank : 53) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| NC score | 0.630438 (rank : 47) | θ value | 9.92553e-07 (rank : 57) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
ATD3A_HUMAN
|
||||||
| NC score | 0.580844 (rank : 48) | θ value | 1.87187e-05 (rank : 71) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.573280 (rank : 49) | θ value | 2.21117e-06 (rank : 61) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.565695 (rank : 50) | θ value | 2.21117e-06 (rank : 60) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.394978 (rank : 51) | θ value | 0.0563607 (rank : 94) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| NC score | 0.392676 (rank : 52) | θ value | 0.0563607 (rank : 95) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.386378 (rank : 53) | θ value | 0.47712 (rank : 101) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
RUVB1_HUMAN
|
||||||
| NC score | 0.285614 (rank : 54) | θ value | 0.47712 (rank : 103) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| NC score | 0.285578 (rank : 55) | θ value | 0.47712 (rank : 104) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
BRD7_HUMAN
|
||||||
| NC score | 0.268944 (rank : 56) | θ value | 1.29631e-06 (rank : 58) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
BRD7_MOUSE
|
||||||
| NC score | 0.264503 (rank : 57) | θ value | 1.29631e-06 (rank : 59) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
BRD9_MOUSE
|
||||||
| NC score | 0.260294 (rank : 58) | θ value | 9.29e-05 (rank : 81) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
BRD9_HUMAN
|
||||||
| NC score | 0.259334 (rank : 59) | θ value | 9.29e-05 (rank : 80) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.258041 (rank : 60) | θ value | 5.44631e-05 (rank : 78) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.257821 (rank : 61) | θ value | 7.59969e-07 (rank : 54) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
BRD3_HUMAN
|
||||||
| NC score | 0.257585 (rank : 62) | θ value | 3.77169e-06 (rank : 63) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
TAF1_HUMAN
|
||||||
| NC score | 0.257247 (rank : 63) | θ value | 7.59969e-07 (rank : 56) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
PCAF_HUMAN
|
||||||
| NC score | 0.253812 (rank : 64) | θ value | 0.00020696 (rank : 83) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92831, Q6NSK1 | Gene names | PCAF | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase PCAF (EC 2.3.1.48) (P300/CBP-associated factor) (P/CAF) (Histone acetylase PCAF). | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.247133 (rank : 65) | θ value | 6.43352e-06 (rank : 67) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.246847 (rank : 66) | θ value | 7.59969e-07 (rank : 55) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
GCNL2_HUMAN
|
||||||
| NC score | 0.246648 (rank : 67) | θ value | 1.43324e-05 (rank : 70) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
BRDT_MOUSE
|
||||||
| NC score | 0.246488 (rank : 68) | θ value | 0.000158464 (rank : 82) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
TAF1L_HUMAN
|
||||||
| NC score | 0.246232 (rank : 69) | θ value | 4.92598e-06 (rank : 65) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.244306 (rank : 70) | θ value | 7.1131e-05 (rank : 79) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.241150 (rank : 71) | θ value | 2.44474e-05 (rank : 74) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.237548 (rank : 72) | θ value | 1.87187e-05 (rank : 72) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.234881 (rank : 73) | θ value | 2.61198e-07 (rank : 50) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.233225 (rank : 74) | θ value | 3.41135e-07 (rank : 51) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.222117 (rank : 75) | θ value | 0.000270298 (rank : 84) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.221181 (rank : 76) | θ value | 1.133e-10 (rank : 44) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.216464 (rank : 77) | θ value | 1.99992e-07 (rank : 49) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.214734 (rank : 78) | θ value | 6.87365e-08 (rank : 48) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.214532 (rank : 79) | θ value | 9.59137e-10 (rank : 46) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.213822 (rank : 80) | θ value | 2.88788e-06 (rank : 62) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.211964 (rank : 81) | θ value | 2.36792 (rank : 128) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.205797 (rank : 82) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.205555 (rank : 83) | θ value | 6.43352e-06 (rank : 66) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.205378 (rank : 84) | θ value | 8.40245e-06 (rank : 69) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.204835 (rank : 85) | θ value | 4.30538e-10 (rank : 45) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.195031 (rank : 86) | θ value | 4.92598e-06 (rank : 64) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.194682 (rank : 87) | θ value | 8.40245e-06 (rank : 68) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.183929 (rank : 88) | θ value | 0.00228821 (rank : 89) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.174839 (rank : 89) | θ value | 0.000461057 (rank : 86) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.174440 (rank : 90) | θ value | 0.0563607 (rank : 93) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.172715 (rank : 91) | θ value | 0.0431538 (rank : 91) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.172228 (rank : 92) | θ value | 0.00035302 (rank : 85) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.148937 (rank : 93) | θ value | 2.36244e-08 (rank : 47) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.148845 (rank : 94) | θ value | 0.163984 (rank : 97) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
BRWD1_HUMAN
|
||||||
| NC score | 0.148756 (rank : 95) | θ value | 4.45536e-07 (rank : 52) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.146307 (rank : 96) | θ value | 0.00102713 (rank : 87) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.143770 (rank : 97) | θ value | 0.00134147 (rank : 88) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.141613 (rank : 98) | θ value | 0.47712 (rank : 105) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.139563 (rank : 99) | θ value | 0.47712 (rank : 106) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.106574 (rank : 100) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.102999 (rank : 101) | θ value | 1.87187e-05 (rank : 73) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.102315 (rank : 102) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RFC4_HUMAN
|
||||||
| NC score | 0.101617 (rank : 103) | θ value | 1.81305 (rank : 122) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RUVB2_HUMAN
|
||||||
| NC score | 0.101304 (rank : 104) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.101129 (rank : 105) | θ value | 2.44474e-05 (rank : 75) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
RUVB2_MOUSE
|
||||||
| NC score | 0.098719 (rank : 106) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
RFC4_MOUSE
|
||||||
| NC score | 0.091670 (rank : 107) | θ value | 1.81305 (rank : 123) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
THNSL_MOUSE
|
||||||
| NC score | 0.090946 (rank : 108) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
THNSL_HUMAN
|
||||||
| NC score | 0.087670 (rank : 109) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.084779 (rank : 110) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.065805 (rank : 111) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
KAD6_MOUSE
|
||||||
| NC score | 0.065482 (rank : 112) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.065115 (rank : 113) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.064102 (rank : 114) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.059542 (rank : 115) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
RFC5_MOUSE
|
||||||
| NC score | 0.056583 (rank : 116) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_HUMAN
|
||||||
| NC score | 0.056061 (rank : 117) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.054925 (rank : 118) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.053970 (rank : 119) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
TIF1B_HUMAN
|
||||||
| NC score | 0.052926 (rank : 120) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TIF1B_MOUSE
|
||||||
| NC score | 0.051889 (rank : 121) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.050523 (rank : 122) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.050277 (rank : 123) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.043106 (rank : 124) | θ value | 0.0736092 (rank : 96) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.042634 (rank : 125) | θ value | 1.38821 (rank : 116) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.040785 (rank : 126) | θ value | 0.0193708 (rank : 90) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.034000 (rank : 127) | θ value | 3.0926 (rank : 132) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.032645 (rank : 128) | θ value | 5.27518 (rank : 146) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.031700 (rank : 129) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.031346 (rank : 130) | θ value | 3.0926 (rank : 129) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.030172 (rank : 131) | θ value | 2.36792 (rank : 126) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
POL2_MOUSE
|
||||||
| NC score | 0.028862 (rank : 132) | θ value | 0.47712 (rank : 102) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11369, Q60713, Q61787 | Gene names | Pol | |||
|
Domain Architecture |

|
|||||
| Description | Retrovirus-related Pol polyprotein LINE-1 (Long interspersed element- 1) (L1) [Includes: Reverse transcriptase (EC 2.7.7.49); Endonuclease]. | |||||
|
CDR2_MOUSE
|
||||||
| NC score | 0.027599 (rank : 133) | θ value | 0.365318 (rank : 100) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97817 | Gene names | Cdr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.026834 (rank : 134) | θ value | 4.03905 (rank : 144) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
LIN1_HUMAN
|
||||||
| NC score | 0.026465 (rank : 135) | θ value | 2.36792 (rank : 127) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08547 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LINE-1 reverse transcriptase homolog. | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.025665 (rank : 136) | θ value | 5.27518 (rank : 151) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.023652 (rank : 137) | θ value | 4.03905 (rank : 141) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
PPHLN_MOUSE
|
||||||
| NC score | 0.023564 (rank : 138) | θ value | 0.62314 (rank : 108) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2H1, Q8BUZ6 | Gene names | Pphln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1. | |||||
|
NAT10_HUMAN
|
||||||
| NC score | 0.022432 (rank : 139) | θ value | 1.06291 (rank : 114) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0A0, Q86WK5, Q9C0F4, Q9HA61, Q9NVF2 | Gene names | NAT10, ALP, KIAA1709 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 10 (EC 2.3.1.-). | |||||
|
CDR2_HUMAN
|
||||||
| NC score | 0.020806 (rank : 140) | θ value | 4.03905 (rank : 137) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01850, Q13977 | Gene names | CDR2, PCD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2 (Paraneoplastic cerebellar degeneration-associated antigen) (Major Yo paraneoplastic antigen). | |||||
|
CF165_HUMAN
|
||||||
| NC score | 0.020584 (rank : 141) | θ value | 1.81305 (rank : 118) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYR0, Q8N9U4 | Gene names | C6orf165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf165. | |||||
|
F10A5_HUMAN
|
||||||
| NC score | 0.020283 (rank : 142) | θ value | 0.279714 (rank : 98) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.019745 (rank : 143) | θ value | 2.36792 (rank : 125) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
NCPR_HUMAN
|
||||||
| NC score | 0.018952 (rank : 144) | θ value | 1.06291 (rank : 115) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16435, Q16455, Q8N181, Q9H3M8 | Gene names | POR, CYPOR | |||
|
Domain Architecture |

|
|||||
| Description | NADPH--cytochrome P450 reductase (EC 1.6.2.4) (CPR) (P450R). | |||||
|
MNDA_HUMAN
|
||||||
| NC score | 0.017500 (rank : 145) | θ value | 0.279714 (rank : 99) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
SETB2_HUMAN
|
||||||
| NC score | 0.016729 (rank : 146) | θ value | 1.81305 (rank : 124) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.016629 (rank : 147) | θ value | 0.0431538 (rank : 92) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.016089 (rank : 148) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYEP_MOUSE
|
||||||
| NC score | 0.015956 (rank : 149) | θ value | 0.813845 (rank : 111) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
COQ9_HUMAN
|
||||||
| NC score | 0.015101 (rank : 150) | θ value | 4.03905 (rank : 138) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75208, Q7L5V7, Q7Z5T6, Q8NBL4, Q9NTJ2, Q9P056 | Gene names | COQ9, C16orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquinone biosynthesis protein COQ9, mitochondrial precursor. | |||||
|
STAR7_MOUSE
|
||||||
| NC score | 0.014033 (rank : 151) | θ value | 3.0926 (rank : 135) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.013330 (rank : 152) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.012261 (rank : 153) | θ value | 3.0926 (rank : 134) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
COQ9_MOUSE
|
||||||
| NC score | 0.012258 (rank : 154) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1Z0, Q9CT61 | Gene names | Coq9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquinone biosynthesis protein COQ9, mitochondrial precursor. | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.012252 (rank : 155) | θ value | 1.38821 (rank : 117) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.011788 (rank : 156) | θ value | 0.62314 (rank : 107) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.010940 (rank : 157) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.010707 (rank : 158) | θ value | 4.03905 (rank : 136) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
JPH4_MOUSE
|
||||||
| NC score | 0.010549 (rank : 159) | θ value | 1.81305 (rank : 120) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
STAR7_HUMAN
|
||||||
| NC score | 0.010543 (rank : 160) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
LCP2_MOUSE
|
||||||
| NC score | 0.010324 (rank : 161) | θ value | 3.0926 (rank : 131) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60787 | Gene names | Lcp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
CT032_HUMAN
|
||||||
| NC score | 0.010136 (rank : 162) | θ value | 4.03905 (rank : 139) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NQ75, Q96K09, Q9BYL5 | Gene names | HEFL, C20orf32 | |||
|
Domain Architecture |

|
|||||
| Description | HEF-like protein. | |||||
|
TF3C4_MOUSE
|
||||||
| NC score | 0.009976 (rank : 163) | θ value | 4.03905 (rank : 143) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BMQ2, Q8BKZ4 | Gene names | Gtf3c4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 4 (EC 2.3.1.48) (Transcription factor IIIC subunit delta) (TF3C-delta) (TFIIIC 90 kDa subunit) (TFIIIC 90). | |||||
|
MRP6_HUMAN
|
||||||
| NC score | 0.009632 (rank : 164) | θ value | 1.06291 (rank : 113) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95255, P78420, Q9UMZ7 | Gene names | ABCC6, ARA, MRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance-associated protein 6 (ATP-binding cassette sub- family C member 6) (Anthracycline resistance-associated protein) (Multi-specific organic anion transporter-E) (MOAT-E). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.009505 (rank : 165) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.009243 (rank : 166) | θ value | 3.0926 (rank : 130) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
DNJC4_MOUSE
|
||||||
| NC score | 0.009089 (rank : 167) | θ value | 1.81305 (rank : 119) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
KIF14_HUMAN
|
||||||
| NC score | 0.008650 (rank : 168) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.008555 (rank : 169) | θ value | 5.27518 (rank : 148) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
JPH4_HUMAN
|
||||||
| NC score | 0.008551 (rank : 170) | θ value | 5.27518 (rank : 147) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.008291 (rank : 171) | θ value | 3.0926 (rank : 133) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
KIF3C_MOUSE
|
||||||
| NC score | 0.007938 (rank : 172) | θ value | 0.813845 (rank : 110) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35066, O35229 | Gene names | Kif3c | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
KIF3C_HUMAN
|
||||||
| NC score | 0.007898 (rank : 173) | θ value | 0.813845 (rank : 109) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.006859 (rank : 174) | θ value | 1.06291 (rank : 112) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
UT14B_MOUSE
|
||||||
| NC score | 0.006679 (rank : 175) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
SYBU_MOUSE
|
||||||
| NC score | 0.006539 (rank : 176) | θ value | 5.27518 (rank : 153) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHS8, Q3TQN7, Q401F3, Q401F4, Q571C1, Q6P1J2, Q80XH0, Q8BHS7, Q8BI27 | Gene names | Sybu, Golsyn, Kiaa1472 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntabulin (Syntaxin-1-binding protein) (Golgi-localized syntaphilin- related protein) (m-Golsyn). | |||||
|
NCOA7_MOUSE
|
||||||
| NC score | 0.006416 (rank : 177) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6DFV7, Q3TAR1, Q3UNB3, Q66K05, Q6ZQM4, Q8BJ15, Q8BJ39 | Gene names | Ncoa7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7. | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.005601 (rank : 178) | θ value | 5.27518 (rank : 149) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
RB15B_MOUSE
|
||||||
| NC score | 0.004388 (rank : 179) | θ value | 5.27518 (rank : 152) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
K1H6_HUMAN
|
||||||
| NC score | 0.004141 (rank : 180) | θ value | 1.81305 (rank : 121) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O76013 | Gene names | KRTHA6, HHA6, HKA6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha6 (Hair keratin, type I Ha6). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.002668 (rank : 181) | θ value | 4.03905 (rank : 140) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.002460 (rank : 182) | θ value | 4.03905 (rank : 142) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
K1C16_MOUSE
|
||||||
| NC score | 0.001802 (rank : 183) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2K1 | Gene names | Krt16, Krt1-16 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.000429 (rank : 184) | θ value | 5.27518 (rank : 150) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.000092 (rank : 185) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | -0.000314 (rank : 186) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
ZN518_HUMAN
|
||||||
| NC score | -0.003973 (rank : 187) | θ value | 4.03905 (rank : 145) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6AHZ1, O15044, Q32MP4 | Gene names | ZNF518, KIAA0335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 518. | |||||