Please be patient as the page loads
|
FIGN_MOUSE
|
||||||
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FIGN_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997032 (rank : 2) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 165 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
SPAST_HUMAN
|
||||||
| θ value | 2.32439e-64 (rank : 3) | NC score | 0.941298 (rank : 3) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 6.76295e-64 (rank : 4) | NC score | 0.939964 (rank : 4) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
KTNA1_HUMAN
|
||||||
| θ value | 1.15895e-47 (rank : 5) | NC score | 0.928323 (rank : 5) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KTNA1_MOUSE
|
||||||
| θ value | 3.72821e-46 (rank : 6) | NC score | 0.926348 (rank : 8) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
KATL1_HUMAN
|
||||||
| θ value | 1.08474e-45 (rank : 7) | NC score | 0.927811 (rank : 6) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_MOUSE
|
||||||
| θ value | 1.41672e-45 (rank : 8) | NC score | 0.927732 (rank : 7) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 3.61106e-41 (rank : 9) | NC score | 0.917124 (rank : 12) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 10) | NC score | 0.917628 (rank : 11) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 6.81017e-40 (rank : 11) | NC score | 0.913986 (rank : 13) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 6.81017e-40 (rank : 12) | NC score | 0.913944 (rank : 14) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
ATAD1_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 13) | NC score | 0.921694 (rank : 9) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| θ value | 1.32909e-35 (rank : 14) | NC score | 0.921694 (rank : 10) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
TERA_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 15) | NC score | 0.873368 (rank : 21) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| θ value | 1.73987e-27 (rank : 16) | NC score | 0.873252 (rank : 22) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PRS8_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 17) | NC score | 0.874216 (rank : 17) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| θ value | 3.87602e-27 (rank : 18) | NC score | 0.874216 (rank : 18) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 2.51237e-26 (rank : 19) | NC score | 0.865628 (rank : 28) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS4_HUMAN
|
||||||
| θ value | 2.51237e-26 (rank : 20) | NC score | 0.871587 (rank : 23) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 21) | NC score | 0.871587 (rank : 24) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS10_HUMAN
|
||||||
| θ value | 9.54697e-26 (rank : 22) | NC score | 0.875185 (rank : 15) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| θ value | 9.54697e-26 (rank : 23) | NC score | 0.875185 (rank : 16) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 24) | NC score | 0.865535 (rank : 29) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS6A_HUMAN
|
||||||
| θ value | 2.77775e-25 (rank : 25) | NC score | 0.873793 (rank : 20) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6A_MOUSE
|
||||||
| θ value | 4.73814e-25 (rank : 26) | NC score | 0.873859 (rank : 19) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 1.99067e-23 (rank : 27) | NC score | 0.826511 (rank : 40) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
PRS6B_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 28) | NC score | 0.868300 (rank : 25) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 29) | NC score | 0.809353 (rank : 41) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
PRS6B_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 30) | NC score | 0.866481 (rank : 26) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
PRS7_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 31) | NC score | 0.865335 (rank : 30) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 32) | NC score | 0.865327 (rank : 31) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 2.43343e-21 (rank : 33) | NC score | 0.856902 (rank : 32) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 3.17815e-21 (rank : 34) | NC score | 0.856532 (rank : 33) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 35) | NC score | 0.866079 (rank : 27) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 36) | NC score | 0.839770 (rank : 34) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
YMEL1_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 37) | NC score | 0.837105 (rank : 36) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
AFG32_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 38) | NC score | 0.829167 (rank : 38) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 39) | NC score | 0.829100 (rank : 39) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
SPG7_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 40) | NC score | 0.837520 (rank : 35) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
AFG31_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 41) | NC score | 0.832934 (rank : 37) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
NSF_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 42) | NC score | 0.802212 (rank : 42) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
NSF_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 43) | NC score | 0.801697 (rank : 43) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 44) | NC score | 0.585677 (rank : 47) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
ATD3A_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 45) | NC score | 0.604984 (rank : 46) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 46) | NC score | 0.040770 (rank : 78) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 47) | NC score | 0.585335 (rank : 48) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
TRP13_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 48) | NC score | 0.659737 (rank : 44) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 49) | NC score | 0.654913 (rank : 45) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 50) | NC score | 0.037218 (rank : 84) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 51) | NC score | 0.058305 (rank : 68) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
DCBD1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 52) | NC score | 0.043077 (rank : 77) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 53) | NC score | 0.050213 (rank : 73) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
TLE3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 54) | NC score | 0.025637 (rank : 102) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04726, Q8IVV6, Q8WVR2, Q9HCM5 | Gene names | TLE3, KIAA1547 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG3). | |||||
|
OTU7B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 55) | NC score | 0.037856 (rank : 81) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6GQQ9, Q8WWA7, Q9NQ53, Q9UFF4 | Gene names | OTUD7B, ZA20D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7B (EC 3.-.-.-) (Zinc finger protein Cezanne) (Zinc finger A20 domain-containing protein 1) (Cellular zinc finger anti-NF-kappa B protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 56) | NC score | 0.032811 (rank : 88) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 57) | NC score | 0.027910 (rank : 96) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
BAG4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 58) | NC score | 0.053709 (rank : 70) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CI61, Q3TRL9, Q91VT5, Q9CWG2 | Gene names | Bag4, Sodd | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 0.163984 (rank : 59) | NC score | 0.033758 (rank : 87) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
PAX7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 60) | NC score | 0.015898 (rank : 127) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23759 | Gene names | PAX7, HUP1 | |||
|
Domain Architecture |
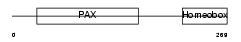
|
|||||
| Description | Paired box protein Pax-7 (HUP1). | |||||
|
RUVB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 61) | NC score | 0.301594 (rank : 54) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 62) | NC score | 0.301556 (rank : 55) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
THNSL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 63) | NC score | 0.138783 (rank : 58) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 0.279714 (rank : 64) | NC score | 0.055569 (rank : 69) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 0.279714 (rank : 65) | NC score | 0.386581 (rank : 51) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| θ value | 0.279714 (rank : 66) | NC score | 0.383491 (rank : 52) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
HXB3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 67) | NC score | 0.014947 (rank : 133) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
FUS_HUMAN
|
||||||
| θ value | 0.365318 (rank : 68) | NC score | 0.034314 (rank : 86) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 69) | NC score | 0.037366 (rank : 82) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 70) | NC score | 0.045416 (rank : 76) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 71) | NC score | 0.039654 (rank : 80) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.027049 (rank : 98) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 73) | NC score | 0.047970 (rank : 74) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PAX3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 74) | NC score | 0.017137 (rank : 124) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |
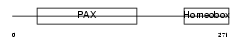
|
|||||
| Description | Paired box protein Pax-3. | |||||
|
THNSL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.126894 (rank : 59) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
TLE3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.022844 (rank : 110) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08122, Q923A4 | Gene names | Tle3, Esg | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG) (Grg-3). | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 0.62314 (rank : 77) | NC score | 0.007491 (rank : 157) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ANC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.028995 (rank : 93) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53995, Q8BP33, Q8C772 | Gene names | Anapc1, Mcpr, Tsg24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
GLIS3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.001785 (rank : 168) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
MAML1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.037008 (rank : 85) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 0.813845 (rank : 81) | NC score | 0.045762 (rank : 75) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
PAX3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 82) | NC score | 0.016557 (rank : 126) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |
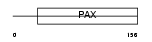
|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
ZF106_MOUSE
|
||||||
| θ value | 0.813845 (rank : 83) | NC score | 0.020049 (rank : 116) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
CTBP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.023552 (rank : 108) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56546, O54855, O88462 | Gene names | Ctbp2 | |||
|
Domain Architecture |
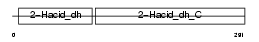
|
|||||
| Description | C-terminal-binding protein 2 (CtBP2). | |||||
|
FUS_MOUSE
|
||||||
| θ value | 1.06291 (rank : 85) | NC score | 0.028557 (rank : 95) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 86) | NC score | 0.028889 (rank : 94) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 87) | NC score | 0.024786 (rank : 103) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 88) | NC score | 0.022191 (rank : 112) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 89) | NC score | 0.029284 (rank : 91) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
LCOR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 90) | NC score | 0.037276 (rank : 83) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZPI3, Q3U302, Q5CZW7, Q80VA8, Q8BGT2, Q8C9Q0 | Gene names | Lcor, Kiaa1795, Mlr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
LEF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 91) | NC score | 0.018678 (rank : 120) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.016764 (rank : 125) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.017434 (rank : 122) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 1.38821 (rank : 94) | NC score | 0.001548 (rank : 169) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.023766 (rank : 107) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.051998 (rank : 71) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
EPOR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.022509 (rank : 111) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19235, Q15443 | Gene names | EPOR | |||
|
Domain Architecture |
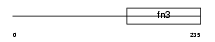
|
|||||
| Description | Erythropoietin receptor precursor (EPO-R). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.029793 (rank : 90) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.020150 (rank : 115) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 100) | NC score | 0.029181 (rank : 92) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 1.81305 (rank : 101) | NC score | 0.012637 (rank : 139) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SP5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 102) | NC score | 0.000573 (rank : 173) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.009124 (rank : 150) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.009019 (rank : 151) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CD248_HUMAN
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.011131 (rank : 146) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.027365 (rank : 97) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
DIP2A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 107) | NC score | 0.015175 (rank : 131) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BWT5, Q8CHI0 | Gene names | Dip2a, Dip2, Kiaa0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
HXB3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 108) | NC score | 0.011545 (rank : 145) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
LONM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 109) | NC score | 0.377627 (rank : 53) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
SP5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 110) | NC score | 0.000704 (rank : 172) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 111) | NC score | 0.019054 (rank : 119) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
AATK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.000966 (rank : 171) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.012629 (rank : 140) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.021060 (rank : 114) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
K1543_HUMAN
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.031341 (rank : 89) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MARK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | -0.004664 (rank : 177) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHJ5, Q69ZI7 | Gene names | Mark1, Emk3, Kiaa1477 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK1 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 1) (ELKL motif serine/threonine-protein kinase 3). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.011945 (rank : 142) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.003322 (rank : 165) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.022878 (rank : 109) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.022000 (rank : 113) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
ELK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.009409 (rank : 149) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
LPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.014049 (rank : 134) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MEF2A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.015898 (rank : 128) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60929 | Gene names | Mef2a | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2A. | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.008679 (rank : 154) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
NFAC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.015271 (rank : 129) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NIBA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.019853 (rank : 118) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
NTRK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | -0.004758 (rank : 178) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15209, Q3TUF9, Q80WU0, Q91XJ9 | Gene names | Ntrk2, Trkb | |||
|
Domain Architecture |

|
|||||
| Description | BDNF/NT-3 growth factors receptor precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase receptor type 2) (TrkB tyrosine kinase) (GP145-TrkB/GP95-TrkB) (Trk-B). | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.006728 (rank : 158) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.025662 (rank : 101) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.011038 (rank : 147) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TGIF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 131) | NC score | 0.006180 (rank : 159) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70284, Q921F9, Q99JS3 | Gene names | Tgif | |||
|
Domain Architecture |
|
|||||
| Description | 5'-TG-3'-interacting factor (Homeobox protein TGIF). | |||||
|
ZN438_HUMAN
|
||||||
| θ value | 4.03905 (rank : 132) | NC score | 0.001411 (rank : 170) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
DIP2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.013366 (rank : 137) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14689, Q8IVA3, Q8N4S2, Q8TD89, Q96ML9 | Gene names | DIP2A, C21orf106, DIP2, KIAA0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.040161 (rank : 79) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
ESAM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.008876 (rank : 153) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AP7, Q96T50 | Gene names | ESAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
LCOR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.026980 (rank : 99) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JN0, Q5VW16, Q7Z723, Q86T32, Q86T33, Q8N3L6 | Gene names | LCOR, KIAA1795, MLR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
LEF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.012156 (rank : 141) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
MUC15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.011883 (rank : 143) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N387, Q6UWS3, Q8IXI8, Q8WW41 | Gene names | MUC15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.018355 (rank : 121) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.004108 (rank : 163) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
PTPRK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.002332 (rank : 167) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
S45A3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.008905 (rank : 152) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K0H7, Q8K252, Q8R1I0 | Gene names | Slc45a3, Pcanap6, Prst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 45 member 3 (Prostate cancer-associated protein 6) (Prostein). | |||||
|
HNRL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.015205 (rank : 130) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.024359 (rank : 104) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
NRG3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.015011 (rank : 132) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P56975 | Gene names | NRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-3, membrane-bound isoform precursor (Pro-NRG3) [Contains: Neuregulin-3 (NRG-3)]. | |||||
|
NUP53_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.017348 (rank : 123) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.023891 (rank : 106) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PRDM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | -0.004039 (rank : 176) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.003083 (rank : 166) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.013657 (rank : 136) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.011620 (rank : 144) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.005546 (rank : 161) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
AF9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.012764 (rank : 138) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
AL2S2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | -0.004017 (rank : 175) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4T3, Q6P922, Q8BU86, Q8BXK0, Q8K4T2 | Gene names | Als2cr2, Papk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ALS2CR2 (EC 2.7.11.1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 2 protein homolog) (ILP-interacting protein homolog) (Polyploidy-associated protein kinase). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.024143 (rank : 105) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.004626 (rank : 162) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.019998 (rank : 117) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CTBP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.013800 (rank : 135) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56545, O43449 | Gene names | CTBP2 | |||
|
Domain Architecture |
|
|||||
| Description | C-terminal-binding protein 2 (CtBP2). | |||||
|
LPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.010493 (rank : 148) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
MYBB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.005667 (rank : 160) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48972 | Gene names | Mybl2, Bmyb | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
NKX26_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.004013 (rank : 164) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43688 | Gene names | Nkx2-6, Nkx-2.6, Nkx2f | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.6 (Homeobox protein NK-2 homolog F). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.007941 (rank : 156) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
TLN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.008272 (rank : 155) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.025820 (rank : 100) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | -0.000559 (rank : 174) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
BCS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.518636 (rank : 49) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.506550 (rank : 50) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
KAD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.092168 (rank : 63) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
KAD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.071440 (rank : 67) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.091778 (rank : 64) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.100854 (rank : 62) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RFC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.082161 (rank : 65) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RFC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.072412 (rank : 66) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
RUVB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.107165 (rank : 60) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.103011 (rank : 61) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
SPG20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.050701 (rank : 72) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R1X6, Q6ZQ87, Q8BJD3, Q8BM37, Q8BZ63 | Gene names | Spg20, Kiaa0610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spartin. | |||||
|
WRIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.192172 (rank : 56) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.191233 (rank : 57) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 165 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.997032 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
SPAST_HUMAN
|
||||||
| NC score | 0.941298 (rank : 3) | θ value | 2.32439e-64 (rank : 3) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.939964 (rank : 4) | θ value | 6.76295e-64 (rank : 4) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
KTNA1_HUMAN
|
||||||
| NC score | 0.928323 (rank : 5) | θ value | 1.15895e-47 (rank : 5) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KATL1_HUMAN
|
||||||
| NC score | 0.927811 (rank : 6) | θ value | 1.08474e-45 (rank : 7) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_MOUSE
|
||||||
| NC score | 0.927732 (rank : 7) | θ value | 1.41672e-45 (rank : 8) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KTNA1_MOUSE
|
||||||
| NC score | 0.926348 (rank : 8) | θ value | 3.72821e-46 (rank : 6) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
ATAD1_HUMAN
|
||||||
| NC score | 0.921694 (rank : 9) | θ value | 1.32909e-35 (rank : 13) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| NC score | 0.921694 (rank : 10) | θ value | 1.32909e-35 (rank : 14) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.917628 (rank : 11) | θ value | 8.04462e-41 (rank : 10) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.917124 (rank : 12) | θ value | 3.61106e-41 (rank : 9) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.913986 (rank : 13) | θ value | 6.81017e-40 (rank : 11) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.913944 (rank : 14) | θ value | 6.81017e-40 (rank : 12) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
PRS10_HUMAN
|
||||||
| NC score | 0.875185 (rank : 15) | θ value | 9.54697e-26 (rank : 22) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| NC score | 0.875185 (rank : 16) | θ value | 9.54697e-26 (rank : 23) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS8_HUMAN
|
||||||
| NC score | 0.874216 (rank : 17) | θ value | 3.87602e-27 (rank : 17) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| NC score | 0.874216 (rank : 18) | θ value | 3.87602e-27 (rank : 18) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS6A_MOUSE
|
||||||
| NC score | 0.873859 (rank : 19) | θ value | 4.73814e-25 (rank : 26) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6A_HUMAN
|
||||||
| NC score | 0.873793 (rank : 20) | θ value | 2.77775e-25 (rank : 25) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
TERA_HUMAN
|
||||||
| NC score | 0.873368 (rank : 21) | θ value | 1.33217e-27 (rank : 15) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| NC score | 0.873252 (rank : 22) | θ value | 1.73987e-27 (rank : 16) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PRS4_HUMAN
|
||||||
| NC score | 0.871587 (rank : 23) | θ value | 2.51237e-26 (rank : 20) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| NC score | 0.871587 (rank : 24) | θ value | 2.51237e-26 (rank : 21) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS6B_HUMAN
|
||||||
| NC score | 0.868300 (rank : 25) | θ value | 3.39556e-23 (rank : 28) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| NC score | 0.866481 (rank : 26) | θ value | 3.75424e-22 (rank : 30) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.866079 (rank : 27) | θ value | 5.42112e-21 (rank : 35) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.865628 (rank : 28) | θ value | 2.51237e-26 (rank : 19) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.865535 (rank : 29) | θ value | 1.24688e-25 (rank : 24) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS7_HUMAN
|
||||||
| NC score | 0.865335 (rank : 30) | θ value | 8.36355e-22 (rank : 31) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| NC score | 0.865327 (rank : 31) | θ value | 8.36355e-22 (rank : 32) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.856902 (rank : 32) | θ value | 2.43343e-21 (rank : 33) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.856532 (rank : 33) | θ value | 3.17815e-21 (rank : 34) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.839770 (rank : 34) | θ value | 9.56915e-18 (rank : 36) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
SPG7_HUMAN
|
||||||
| NC score | 0.837520 (rank : 35) | θ value | 4.74913e-17 (rank : 40) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
YMEL1_HUMAN
|
||||||
| NC score | 0.837105 (rank : 36) | θ value | 1.63225e-17 (rank : 37) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
AFG31_MOUSE
|
||||||
| NC score | 0.832934 (rank : 37) | θ value | 1.058e-16 (rank : 41) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
AFG32_HUMAN
|
||||||
| NC score | 0.829167 (rank : 38) | θ value | 2.7842e-17 (rank : 38) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| NC score | 0.829100 (rank : 39) | θ value | 2.7842e-17 (rank : 39) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.826511 (rank : 40) | θ value | 1.99067e-23 (rank : 27) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.809353 (rank : 41) | θ value | 1.6852e-22 (rank : 29) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
NSF_HUMAN
|
||||||
| NC score | 0.802212 (rank : 42) | θ value | 2.79066e-09 (rank : 42) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
NSF_MOUSE
|
||||||
| NC score | 0.801697 (rank : 43) | θ value | 2.79066e-09 (rank : 43) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
TRP13_MOUSE
|
||||||
| NC score | 0.659737 (rank : 44) | θ value | 0.000121331 (rank : 48) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| NC score | 0.654913 (rank : 45) | θ value | 0.000461057 (rank : 49) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
ATD3A_HUMAN
|
||||||
| NC score | 0.604984 (rank : 46) | θ value | 3.77169e-06 (rank : 45) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.585677 (rank : 47) | θ value | 2.61198e-07 (rank : 44) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.585335 (rank : 48) | θ value | 2.44474e-05 (rank : 47) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
BCS1_HUMAN
|
||||||
| NC score | 0.518636 (rank : 49) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| NC score | 0.506550 (rank : 50) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.386581 (rank : 51) | θ value | 0.279714 (rank : 65) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| NC score | 0.383491 (rank : 52) | θ value | 0.279714 (rank : 66) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.377627 (rank : 53) | θ value | 2.36792 (rank : 109) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
RUVB1_HUMAN
|
||||||
| NC score | 0.301594 (rank : 54) | θ value | 0.21417 (rank : 61) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| NC score | 0.301556 (rank : 55) | θ value | 0.21417 (rank : 62) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.192172 (rank : 56) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.191233 (rank : 57) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
THNSL_MOUSE
|
||||||
| NC score | 0.138783 (rank : 58) | θ value | 0.21417 (rank : 63) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
THNSL_HUMAN
|
||||||
| NC score | 0.126894 (rank : 59) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
RUVB2_HUMAN
|
||||||
| NC score | 0.107165 (rank : 60) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| NC score | 0.103011 (rank : 61) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.100854 (rank : 62) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.092168 (rank : 63) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.091778 (rank : 64) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RFC4_HUMAN
|
||||||
| NC score | 0.082161 (rank : 65) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RFC4_MOUSE
|
||||||
| NC score | 0.072412 (rank : 66) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
KAD6_MOUSE
|
||||||
| NC score | 0.071440 (rank : 67) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.058305 (rank : 68) | θ value | 0.00869519 (rank : 51) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.055569 (rank : 69) | θ value | 0.279714 (rank : 64) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
BAG4_MOUSE
|
||||||
| NC score | 0.053709 (rank : 70) | θ value | 0.163984 (rank : 58) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CI61, Q3TRL9, Q91VT5, Q9CWG2 | Gene names | Bag4, Sodd | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.051998 (rank : 71) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
SPG20_MOUSE
|
||||||
| NC score | 0.050701 (rank : 72) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R1X6, Q6ZQ87, Q8BJD3, Q8BM37, Q8BZ63 | Gene names | Spg20, Kiaa0610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spartin. | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.050213 (rank : 73) | θ value | 0.0252991 (rank : 53) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.047970 (rank : 74) | θ value | 0.62314 (rank : 73) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.045762 (rank : 75) | θ value | 0.813845 (rank : 81) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.045416 (rank : 76) | θ value | 0.47712 (rank : 70) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
DCBD1_MOUSE
|
||||||
| NC score | 0.043077 (rank : 77) | θ value | 0.0252991 (rank : 52) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.040770 (rank : 78) | θ value | 6.43352e-06 (rank : 46) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.040161 (rank : 79) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.039654 (rank : 80) | θ value | 0.47712 (rank : 71) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
OTU7B_HUMAN
|
||||||
| NC score | 0.037856 (rank : 81) | θ value | 0.0563607 (rank : 55) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6GQQ9, Q8WWA7, Q9NQ53, Q9UFF4 | Gene names | OTUD7B, ZA20D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7B (EC 3.-.-.-) (Zinc finger protein Cezanne) (Zinc finger A20 domain-containing protein 1) (Cellular zinc finger anti-NF-kappa B protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.037366 (rank : 82) | θ value | 0.365318 (rank : 69) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
LCOR_MOUSE
|
||||||
| NC score | 0.037276 (rank : 83) | θ value | 1.38821 (rank : 90) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZPI3, Q3U302, Q5CZW7, Q80VA8, Q8BGT2, Q8C9Q0 | Gene names | Lcor, Kiaa1795, Mlr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.037218 (rank : 84) | θ value | 0.00390308 (rank : 50) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MAML1_MOUSE
|
||||||
| NC score | 0.037008 (rank : 85) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.034314 (rank : 86) | θ value | 0.365318 (rank : 68) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.033758 (rank : 87) | θ value | 0.163984 (rank : 59) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.032811 (rank : 88) | θ value | 0.0563607 (rank : 56) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.031341 (rank : 89) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.029793 (rank : 90) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.029284 (rank : 91) | θ value | 1.06291 (rank : 89) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.029181 (rank : 92) | θ value | 1.81305 (rank : 100) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
ANC1_MOUSE
|
||||||
| NC score | 0.028995 (rank : 93) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53995, Q8BP33, Q8C772 | Gene names | Anapc1, Mcpr, Tsg24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.028889 (rank : 94) | θ value | 1.06291 (rank : 86) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.028557 (rank : 95) | θ value | 1.06291 (rank : 85) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.027910 (rank : 96) | θ value | 0.0961366 (rank : 57) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.027365 (rank : 97) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.027049 (rank : 98) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
LCOR_HUMAN
|
||||||
| NC score | 0.026980 (rank : 99) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JN0, Q5VW16, Q7Z723, Q86T32, Q86T33, Q8N3L6 | Gene names | LCOR, KIAA1795, MLR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.025820 (rank : 100) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.025662 (rank : 101) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
TLE3_HUMAN
|
||||||
| NC score | 0.025637 (rank : 102) | θ value | 0.0431538 (rank : 54) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04726, Q8IVV6, Q8WVR2, Q9HCM5 | Gene names | TLE3, KIAA1547 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG3). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.024786 (rank : 103) | θ value | 1.06291 (rank : 87) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.024359 (rank : 104) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.024143 (rank : 105) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.023891 (rank : 106) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.023766 (rank : 107) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CTBP2_MOUSE
|
||||||
| NC score | 0.023552 (rank : 108) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56546, O54855, O88462 | Gene names | Ctbp2 | |||
|
Domain Architecture |

|
|||||
| Description | C-terminal-binding protein 2 (CtBP2). | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.022878 (rank : 109) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
TLE3_MOUSE
|
||||||
| NC score | 0.022844 (rank : 110) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08122, Q923A4 | Gene names | Tle3, Esg | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG) (Grg-3). | |||||
|
EPOR_HUMAN
|
||||||
| NC score | 0.022509 (rank : 111) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19235, Q15443 | Gene names | EPOR | |||
|
Domain Architecture |

|
|||||
| Description | Erythropoietin receptor precursor (EPO-R). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.022191 (rank : 112) | θ value | 1.06291 (rank : 88) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.022000 (rank : 113) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.021060 (rank : 114) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.020150 (rank : 115) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
ZF106_MOUSE
|
||||||
| NC score | 0.020049 (rank : 116) | θ value | 0.813845 (rank : 83) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.019998 (rank : 117) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NIBA_HUMAN
|
||||||
| NC score | 0.019853 (rank : 118) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.019054 (rank : 119) | θ value | 2.36792 (rank : 111) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
LEF1_MOUSE
|
||||||
| NC score | 0.018678 (rank : 120) | θ value | 1.38821 (rank : 91) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.018355 (rank : 121) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.017434 (rank : 122) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
NUP53_HUMAN
|
||||||
| NC score | 0.017348 (rank : 123) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
PAX3_MOUSE
|
||||||
| NC score | 0.017137 (rank : 124) | θ value | 0.62314 (rank : 74) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.016764 (rank : 125) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
PAX3_HUMAN
|
||||||
| NC score | 0.016557 (rank : 126) | θ value | 0.813845 (rank : 82) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
PAX7_HUMAN
|
||||||
| NC score | 0.015898 (rank : 127) | θ value | 0.21417 (rank : 60) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23759 | Gene names | PAX7, HUP1 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-7 (HUP1). | |||||
|
MEF2A_MOUSE
|
||||||
| NC score | 0.015898 (rank : 128) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60929 | Gene names | Mef2a | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2A. | |||||
|
NFAC2_HUMAN
|
||||||
| NC score | 0.015271 (rank : 129) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
HNRL1_MOUSE
|
||||||
| NC score | 0.015205 (rank : 130) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
DIP2A_MOUSE
|
||||||
| NC score | 0.015175 (rank : 131) | θ value | 2.36792 (rank : 107) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BWT5, Q8CHI0 | Gene names | Dip2a, Dip2, Kiaa0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
NRG3_HUMAN
|
||||||
| NC score | 0.015011 (rank : 132) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P56975 | Gene names | NRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-3, membrane-bound isoform precursor (Pro-NRG3) [Contains: Neuregulin-3 (NRG-3)]. | |||||
|
HXB3_HUMAN
|
||||||
| NC score | 0.014947 (rank : 133) | θ value | 0.279714 (rank : 67) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.014049 (rank : 134) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
CTBP2_HUMAN
|
||||||
| NC score | 0.013800 (rank : 135) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56545, O43449 | Gene names | CTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-terminal-binding protein 2 (CtBP2). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.013657 (rank : 136) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
DIP2A_HUMAN
|
||||||
| NC score | 0.013366 (rank : 137) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14689, Q8IVA3, Q8N4S2, Q8TD89, Q96ML9 | Gene names | DIP2A, C21orf106, DIP2, KIAA0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.012764 (rank : 138) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.012637 (rank : 139) | θ value | 1.81305 (rank : 101) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.012629 (rank : 140) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
LEF1_HUMAN
|
||||||
| NC score | 0.012156 (rank : 141) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.011945 (rank : 142) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
MUC15_HUMAN
|
||||||
| NC score | 0.011883 (rank : 143) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N387, Q6UWS3, Q8IXI8, Q8WW41 | Gene names | MUC15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.011620 (rank : 144) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
HXB3_MOUSE
|
||||||
| NC score | 0.011545 (rank : 145) | θ value | 2.36792 (rank : 108) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.011131 (rank : 146) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.011038 (rank : 147) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
LPP_MOUSE
|
||||||
| NC score | 0.010493 (rank : 148) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.009409 (rank : 149) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.009124 (rank : 150) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.009019 (rank : 151) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
S45A3_MOUSE
|
||||||
| NC score | 0.008905 (rank : 152) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K0H7, Q8K252, Q8R1I0 | Gene names | Slc45a3, Pcanap6, Prst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 45 member 3 (Prostate cancer-associated protein 6) (Prostein). | |||||
|
ESAM_HUMAN
|
||||||
| NC score | 0.008876 (rank : 153) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AP7, Q96T50 | Gene names | ESAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.008679 (rank : 154) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.008272 (rank : 155) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.007941 (rank : 156) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.007491 (rank : 157) | θ value | 0.62314 (rank : 77) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.006728 (rank : 158) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
TGIF_MOUSE
|
||||||
| NC score | 0.006180 (rank : 159) | θ value | 4.03905 (rank : 131) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70284, Q921F9, Q99JS3 | Gene names | Tgif | |||
|
Domain Architecture |

|
|||||
| Description | 5'-TG-3'-interacting factor (Homeobox protein TGIF). | |||||
|
MYBB_MOUSE
|
||||||
| NC score | 0.005667 (rank : 160) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48972 | Gene names | Mybl2, Bmyb | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.005546 (rank : 161) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.004626 (rank : 162) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.004108 (rank : 163) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
NKX26_MOUSE
|
||||||
| NC score | 0.004013 (rank : 164) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43688 | Gene names | Nkx2-6, Nkx-2.6, Nkx2f | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.6 (Homeobox protein NK-2 homolog F). | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.003322 (rank : 165) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.003083 (rank : 166) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
PTPRK_HUMAN
|
||||||
| NC score | 0.002332 (rank : 167) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
GLIS3_MOUSE
|
||||||
| NC score | 0.001785 (rank : 168) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.001548 (rank : 169) | θ value | 1.38821 (rank : 94) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
ZN438_HUMAN
|
||||||
| NC score | 0.001411 (rank : 170) | θ value | 4.03905 (rank : 132) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.000966 (rank : 171) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.000704 (rank : 172) | θ value | 2.36792 (rank : 110) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
SP5_HUMAN
|
||||||
| NC score | 0.000573 (rank : 173) | θ value | 1.81305 (rank : 102) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | -0.000559 (rank : 174) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
AL2S2_MOUSE
|
||||||
| NC score | -0.004017 (rank : 175) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4T3, Q6P922, Q8BU86, Q8BXK0, Q8K4T2 | Gene names | Als2cr2, Papk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ALS2CR2 (EC 2.7.11.1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 2 protein homolog) (ILP-interacting protein homolog) (Polyploidy-associated protein kinase). | |||||
|
PRDM1_MOUSE
|
||||||
| NC score | -0.004039 (rank : 176) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||
|
MARK1_MOUSE
|
||||||
| NC score | -0.004664 (rank : 177) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHJ5, Q69ZI7 | Gene names | Mark1, Emk3, Kiaa1477 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK1 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 1) (ELKL motif serine/threonine-protein kinase 3). | |||||
|
NTRK2_MOUSE
|
||||||
| NC score | -0.004758 (rank : 178) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 165 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15209, Q3TUF9, Q80WU0, Q91XJ9 | Gene names | Ntrk2, Trkb | |||
|
Domain Architecture |

|
|||||
| Description | BDNF/NT-3 growth factors receptor precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase receptor type 2) (TrkB tyrosine kinase) (GP145-TrkB/GP95-TrkB) (Trk-B). | |||||