Please be patient as the page loads
|
LCOR_HUMAN
|
||||||
| SwissProt Accessions | Q96JN0, Q5VW16, Q7Z723, Q86T32, Q86T33, Q8N3L6 | Gene names | LCOR, KIAA1795, MLR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LCOR_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96JN0, Q5VW16, Q7Z723, Q86T32, Q86T33, Q8N3L6 | Gene names | LCOR, KIAA1795, MLR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
LCOR_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995138 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6ZPI3, Q3U302, Q5CZW7, Q80VA8, Q8BGT2, Q8C9Q0 | Gene names | Lcor, Kiaa1795, Mlr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
MEF2A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 3) | NC score | 0.062366 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02078, O43814, Q14223, Q14224, Q96D14 | Gene names | MEF2A, MEF2 | |||
|
Domain Architecture |
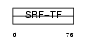
|
|||||
| Description | Myocyte-specific enhancer factor 2A (Serum response factor-like protein 1). | |||||
|
ELOA3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 4) | NC score | 0.077744 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
GGN_MOUSE
|
||||||
| θ value | 0.47712 (rank : 5) | NC score | 0.062927 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 6) | NC score | 0.048620 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
FGD3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.020806 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 8) | NC score | 0.072761 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.072761 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.045230 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.028756 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
TRM6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.057913 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CE96, Q3TJZ8, Q3TME7, Q6ZPW8, Q80Y59, Q8CEU0 | Gene names | Trm6, Kiaa1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.035624 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
PDPK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.005190 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15530 | Gene names | PDPK1, PDK1 | |||
|
Domain Architecture |
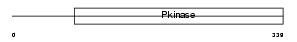
|
|||||
| Description | 3-phosphoinositide-dependent protein kinase 1 (EC 2.7.11.1) (hPDK1). | |||||
|
CBX4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.033085 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.019948 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
PCDA9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.005034 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
C1QB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.018169 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02746, Q5T959, Q96H17 | Gene names | C1QB | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1q subcomponent subunit B precursor. | |||||
|
CO1A2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.022047 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.029109 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
K1024_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.032046 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPX6 | Gene names | KIAA1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024. | |||||
|
PIAS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.022815 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75928, O75927, Q96BT5, Q96KE3 | Gene names | PIAS2, PIASX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (Miz1) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3) (PIAS-NY protein). | |||||
|
PIAS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.022848 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C5D8, O54987, Q8C384, Q8CDQ8, Q8K208, Q99JX5, Q9D5W7, Q9QZ63 | Gene names | Pias2, Miz1, Piasx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3). | |||||
|
SIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.007149 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60315, Q9UED1 | Gene names | ZFHX1B, KIAA0569, SIP1, ZFX1B | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1) (SMADIP1). | |||||
|
WAPL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.048868 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z5K2, Q5VSK5, Q8IX10, Q92549 | Gene names | WAPAL, FOE, KIAA0261, WAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Friend of EBNA2 protein). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.028943 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CO5A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.022760 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CO5A1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.022913 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.026980 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.028337 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
WAPL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.042662 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
CJ119_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.041418 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BTE3, Q6IA56, Q9BVT9, Q9H916 | Gene names | C10orf119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf119. | |||||
|
ELOA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.036223 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
GP111_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.017495 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZF7, Q86SL6, Q8NGU5, Q8TDT5 | Gene names | GPR111, PGR20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 111 (G-protein coupled receptor PGR20). | |||||
|
CJ119_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.038027 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3C0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf119 homolog. | |||||
|
CO4A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.013761 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.012829 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
HME1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.004197 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05925 | Gene names | EN1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein engrailed-1 (Hu-En-1). | |||||
|
IRF8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.013503 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23611 | Gene names | Irf8, Icsbp, Icsbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 8 (IRF-8) (Interferon consensus sequence- binding protein) (ICSBP). | |||||
|
KLK13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.002380 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKR3, Q9Y433 | Gene names | KLK13, KLKL4 | |||
|
Domain Architecture |
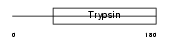
|
|||||
| Description | Kallikrein-13 precursor (EC 3.4.21.-) (Kallikrein-like protein 4) (KLK-L4). | |||||
|
NOL4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.012928 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60954 | Gene names | Nol4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 4. | |||||
|
PLD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.017415 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z280, O35911 | Gene names | Pld1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase D1 (EC 3.1.4.4) (PLD 1) (Choline phosphatase 1) (Phosphatidylcholine-hydrolyzing phospholipase D1) (mPLD1). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.021751 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRDM8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.019881 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQV8, Q6IQ36 | Gene names | PRDM8, PFM5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 8 (PR domain-containing protein 8). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.021158 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
UBP53_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.021714 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
LCOR_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96JN0, Q5VW16, Q7Z723, Q86T32, Q86T33, Q8N3L6 | Gene names | LCOR, KIAA1795, MLR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
LCOR_MOUSE
|
||||||
| NC score | 0.995138 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6ZPI3, Q3U302, Q5CZW7, Q80VA8, Q8BGT2, Q8C9Q0 | Gene names | Lcor, Kiaa1795, Mlr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
ELOA3_HUMAN
|
||||||
| NC score | 0.077744 (rank : 3) | θ value | 0.279714 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.072761 (rank : 4) | θ value | 0.813845 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.072761 (rank : 5) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.062927 (rank : 6) | θ value | 0.47712 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
MEF2A_HUMAN
|
||||||
| NC score | 0.062366 (rank : 7) | θ value | 0.21417 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02078, O43814, Q14223, Q14224, Q96D14 | Gene names | MEF2A, MEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2A (Serum response factor-like protein 1). | |||||
|
TRM6_MOUSE
|
||||||
| NC score | 0.057913 (rank : 8) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CE96, Q3TJZ8, Q3TME7, Q6ZPW8, Q80Y59, Q8CEU0 | Gene names | Trm6, Kiaa1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
WAPL_HUMAN
|
||||||
| NC score | 0.048868 (rank : 9) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z5K2, Q5VSK5, Q8IX10, Q92549 | Gene names | WAPAL, FOE, KIAA0261, WAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Friend of EBNA2 protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.048620 (rank : 10) | θ value | 0.47712 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.045230 (rank : 11) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
WAPL_MOUSE
|
||||||
| NC score | 0.042662 (rank : 12) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
CJ119_HUMAN
|
||||||
| NC score | 0.041418 (rank : 13) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BTE3, Q6IA56, Q9BVT9, Q9H916 | Gene names | C10orf119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf119. | |||||
|
CJ119_MOUSE
|
||||||
| NC score | 0.038027 (rank : 14) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3C0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf119 homolog. | |||||
|
ELOA2_HUMAN
|
||||||
| NC score | 0.036223 (rank : 15) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.035624 (rank : 16) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CBX4_MOUSE
|
||||||
| NC score | 0.033085 (rank : 17) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
K1024_HUMAN
|
||||||
| NC score | 0.032046 (rank : 18) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPX6 | Gene names | KIAA1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024. | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.029109 (rank : 19) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.028943 (rank : 20) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.028756 (rank : 21) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.028337 (rank : 22) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.026980 (rank : 23) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
CO5A1_MOUSE
|
||||||
| NC score | 0.022913 (rank : 24) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
PIAS2_MOUSE
|
||||||
| NC score | 0.022848 (rank : 25) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C5D8, O54987, Q8C384, Q8CDQ8, Q8K208, Q99JX5, Q9D5W7, Q9QZ63 | Gene names | Pias2, Miz1, Piasx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3). | |||||
|
PIAS2_HUMAN
|
||||||
| NC score | 0.022815 (rank : 26) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75928, O75927, Q96BT5, Q96KE3 | Gene names | PIAS2, PIASX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (Miz1) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3) (PIAS-NY protein). | |||||
|
CO5A1_HUMAN
|
||||||
| NC score | 0.022760 (rank : 27) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CO1A2_HUMAN
|
||||||
| NC score | 0.022047 (rank : 28) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.021751 (rank : 29) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
UBP53_MOUSE
|
||||||
| NC score | 0.021714 (rank : 30) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.021158 (rank : 31) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
FGD3_HUMAN
|
||||||
| NC score | 0.020806 (rank : 32) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.019948 (rank : 33) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
PRDM8_HUMAN
|
||||||
| NC score | 0.019881 (rank : 34) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQV8, Q6IQ36 | Gene names | PRDM8, PFM5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 8 (PR domain-containing protein 8). | |||||
|
C1QB_HUMAN
|
||||||
| NC score | 0.018169 (rank : 35) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02746, Q5T959, Q96H17 | Gene names | C1QB | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1q subcomponent subunit B precursor. | |||||
|
GP111_HUMAN
|
||||||
| NC score | 0.017495 (rank : 36) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZF7, Q86SL6, Q8NGU5, Q8TDT5 | Gene names | GPR111, PGR20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 111 (G-protein coupled receptor PGR20). | |||||
|
PLD1_MOUSE
|
||||||
| NC score | 0.017415 (rank : 37) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z280, O35911 | Gene names | Pld1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase D1 (EC 3.1.4.4) (PLD 1) (Choline phosphatase 1) (Phosphatidylcholine-hydrolyzing phospholipase D1) (mPLD1). | |||||
|
CO4A1_MOUSE
|
||||||
| NC score | 0.013761 (rank : 38) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
IRF8_MOUSE
|
||||||
| NC score | 0.013503 (rank : 39) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23611 | Gene names | Irf8, Icsbp, Icsbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 8 (IRF-8) (Interferon consensus sequence- binding protein) (ICSBP). | |||||
|
NOL4_MOUSE
|
||||||
| NC score | 0.012928 (rank : 40) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60954 | Gene names | Nol4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 4. | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.012829 (rank : 41) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
SIP1_HUMAN
|
||||||
| NC score | 0.007149 (rank : 42) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60315, Q9UED1 | Gene names | ZFHX1B, KIAA0569, SIP1, ZFX1B | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1) (SMADIP1). | |||||
|
PDPK1_HUMAN
|
||||||
| NC score | 0.005190 (rank : 43) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15530 | Gene names | PDPK1, PDK1 | |||
|
Domain Architecture |

|
|||||
| Description | 3-phosphoinositide-dependent protein kinase 1 (EC 2.7.11.1) (hPDK1). | |||||
|
PCDA9_HUMAN
|
||||||
| NC score | 0.005034 (rank : 44) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
HME1_HUMAN
|
||||||
| NC score | 0.004197 (rank : 45) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05925 | Gene names | EN1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein engrailed-1 (Hu-En-1). | |||||
|
KLK13_HUMAN
|
||||||
| NC score | 0.002380 (rank : 46) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKR3, Q9Y433 | Gene names | KLK13, KLKL4 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-13 precursor (EC 3.4.21.-) (Kallikrein-like protein 4) (KLK-L4). | |||||