Please be patient as the page loads
|
PRS4_HUMAN
|
||||||
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PRS4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS6B_HUMAN
|
||||||
| θ value | 3.0981e-109 (rank : 3) | NC score | 0.991012 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| θ value | 1.87955e-106 (rank : 4) | NC score | 0.990611 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
PRS6A_HUMAN
|
||||||
| θ value | 3.32542e-95 (rank : 5) | NC score | 0.988391 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS8_HUMAN
|
||||||
| θ value | 3.32542e-95 (rank : 6) | NC score | 0.989284 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| θ value | 3.32542e-95 (rank : 7) | NC score | 0.989284 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS6A_MOUSE
|
||||||
| θ value | 4.34313e-95 (rank : 8) | NC score | 0.988567 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS7_HUMAN
|
||||||
| θ value | 1.40038e-85 (rank : 9) | NC score | 0.987690 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| θ value | 1.40038e-85 (rank : 10) | NC score | 0.987679 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS10_HUMAN
|
||||||
| θ value | 3.81361e-83 (rank : 11) | NC score | 0.987233 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| θ value | 3.81361e-83 (rank : 12) | NC score | 0.987233 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
TERA_HUMAN
|
||||||
| θ value | 1.15626e-55 (rank : 13) | NC score | 0.953555 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| θ value | 1.51013e-55 (rank : 14) | NC score | 0.953504 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
AFG31_MOUSE
|
||||||
| θ value | 2.49495e-50 (rank : 15) | NC score | 0.946937 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
AFG32_HUMAN
|
||||||
| θ value | 3.2585e-50 (rank : 16) | NC score | 0.943359 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| θ value | 5.55816e-50 (rank : 17) | NC score | 0.943085 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 1.51363e-47 (rank : 18) | NC score | 0.880839 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 1.28137e-46 (rank : 19) | NC score | 0.897974 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
SPG7_HUMAN
|
||||||
| θ value | 1.12253e-42 (rank : 20) | NC score | 0.945552 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 2.50074e-42 (rank : 21) | NC score | 0.940482 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 4.2656e-42 (rank : 22) | NC score | 0.939983 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 9.50281e-42 (rank : 23) | NC score | 0.948081 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 1.2411e-41 (rank : 24) | NC score | 0.934074 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 25) | NC score | 0.934167 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
KTNA1_HUMAN
|
||||||
| θ value | 1.51715e-39 (rank : 26) | NC score | 0.923488 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KTNA1_MOUSE
|
||||||
| θ value | 1.98146e-39 (rank : 27) | NC score | 0.921292 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | 1.98146e-39 (rank : 28) | NC score | 0.936398 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
YMEL1_HUMAN
|
||||||
| θ value | 2.58786e-39 (rank : 29) | NC score | 0.932334 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
KATL1_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 30) | NC score | 0.925919 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_MOUSE
|
||||||
| θ value | 4.88048e-38 (rank : 31) | NC score | 0.925821 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
NSF_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 32) | NC score | 0.925002 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
NSF_HUMAN
|
||||||
| θ value | 5.584e-34 (rank : 33) | NC score | 0.924363 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
SPAST_HUMAN
|
||||||
| θ value | 7.29293e-34 (rank : 34) | NC score | 0.917842 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 2.12192e-33 (rank : 35) | NC score | 0.916139 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 4.72714e-33 (rank : 36) | NC score | 0.907779 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 4.72714e-33 (rank : 37) | NC score | 0.907760 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 2.34606e-32 (rank : 38) | NC score | 0.909757 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 39) | NC score | 0.909976 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
ATAD1_HUMAN
|
||||||
| θ value | 6.82597e-32 (rank : 40) | NC score | 0.938061 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| θ value | 6.82597e-32 (rank : 41) | NC score | 0.938061 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 42) | NC score | 0.871587 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 43) | NC score | 0.866006 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
BCS1_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 44) | NC score | 0.723579 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 45) | NC score | 0.715340 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
TRP13_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 46) | NC score | 0.713040 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 47) | NC score | 0.620855 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
TRP13_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 48) | NC score | 0.709378 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 49) | NC score | 0.612743 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
ATD3A_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 50) | NC score | 0.630170 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 51) | NC score | 0.444497 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 52) | NC score | 0.442680 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 53) | NC score | 0.262689 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
LONM_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 54) | NC score | 0.435324 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
WRIP1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 55) | NC score | 0.258302 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
RUVB1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 56) | NC score | 0.318976 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 57) | NC score | 0.318937 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
KAD2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 58) | NC score | 0.037280 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54819, Q16856, Q5TIF7, Q8TCY2, Q8TCY3 | Gene names | AK2, ADK2 | |||
|
Domain Architecture |
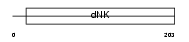
|
|||||
| Description | Adenylate kinase isoenzyme 2, mitochondrial (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
ICA69_HUMAN
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.022340 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q05084, P78506, Q13824 | Gene names | ICA1 | |||
|
Domain Architecture |
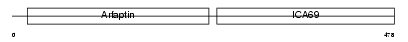
|
|||||
| Description | Islet cell autoantigen 1 (69 kDa islet cell autoantigen) (ICA69) (p69) (Islet cell autoantigen p69) (ICAp69). | |||||
|
RFC5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.078006 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.079422 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
CKAP5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.023277 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14008, Q14668 | Gene names | CKAP5, KIAA0097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 5 (Colonic and hepatic tumor over- expressed protein) (Ch-TOG protein). | |||||
|
LC7L2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.013786 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
LC7L2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.013591 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.001797 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.010465 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
CDC27_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.011458 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
NCOA7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.010810 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NI08, Q3LID6, Q4G0V1, Q5TF95, Q6IPQ4, Q6NE83, Q86T89, Q8N1W4 | Gene names | NCOA7, ERAP140, ESNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7 (140 kDa estrogen receptor-associated protein) (Estrogen nuclear receptor coactivator 1). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.038758 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
RENT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.026915 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.001860 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
IF140_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.012195 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RY7, O60332 | Gene names | IFT140, KIAA0590, WDTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 140 homolog (WD and tetratricopeptide repeats protein 2). | |||||
|
PALM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.009454 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
RENT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.025380 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
RUVB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.122403 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.119852 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
KAD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.018633 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTP6, Q3TKI6, Q9CY37 | Gene names | Ak2, Ak-2 | |||
|
Domain Architecture |
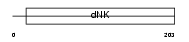
|
|||||
| Description | Adenylate kinase isoenzyme 2, mitochondrial (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | -0.001374 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
THNSL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.104392 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
KAD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.093553 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
P55G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.000590 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.101199 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.114273 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
THNSL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.106712 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
KAD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.071031 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.101004 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RFC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.089972 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
PRS4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS6B_HUMAN
|
||||||
| NC score | 0.991012 (rank : 3) | θ value | 3.0981e-109 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| NC score | 0.990611 (rank : 4) | θ value | 1.87955e-106 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
PRS8_HUMAN
|
||||||
| NC score | 0.989284 (rank : 5) | θ value | 3.32542e-95 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| NC score | 0.989284 (rank : 6) | θ value | 3.32542e-95 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS6A_MOUSE
|
||||||
| NC score | 0.988567 (rank : 7) | θ value | 4.34313e-95 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6A_HUMAN
|
||||||
| NC score | 0.988391 (rank : 8) | θ value | 3.32542e-95 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS7_HUMAN
|
||||||
| NC score | 0.987690 (rank : 9) | θ value | 1.40038e-85 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| NC score | 0.987679 (rank : 10) | θ value | 1.40038e-85 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS10_HUMAN
|
||||||
| NC score | 0.987233 (rank : 11) | θ value | 3.81361e-83 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| NC score | 0.987233 (rank : 12) | θ value | 3.81361e-83 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
TERA_HUMAN
|
||||||
| NC score | 0.953555 (rank : 13) | θ value | 1.15626e-55 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| NC score | 0.953504 (rank : 14) | θ value | 1.51013e-55 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.948081 (rank : 15) | θ value | 9.50281e-42 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
AFG31_MOUSE
|
||||||
| NC score | 0.946937 (rank : 16) | θ value | 2.49495e-50 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
SPG7_HUMAN
|
||||||
| NC score | 0.945552 (rank : 17) | θ value | 1.12253e-42 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
AFG32_HUMAN
|
||||||
| NC score | 0.943359 (rank : 18) | θ value | 3.2585e-50 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| NC score | 0.943085 (rank : 19) | θ value | 5.55816e-50 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.940482 (rank : 20) | θ value | 2.50074e-42 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.939983 (rank : 21) | θ value | 4.2656e-42 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
ATAD1_HUMAN
|
||||||
| NC score | 0.938061 (rank : 22) | θ value | 6.82597e-32 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| NC score | 0.938061 (rank : 23) | θ value | 6.82597e-32 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.936398 (rank : 24) | θ value | 1.98146e-39 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.934167 (rank : 25) | θ value | 2.11701e-41 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.934074 (rank : 26) | θ value | 1.2411e-41 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
YMEL1_HUMAN
|
||||||
| NC score | 0.932334 (rank : 27) | θ value | 2.58786e-39 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
KATL1_HUMAN
|
||||||
| NC score | 0.925919 (rank : 28) | θ value | 4.88048e-38 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_MOUSE
|
||||||
| NC score | 0.925821 (rank : 29) | θ value | 4.88048e-38 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
NSF_MOUSE
|
||||||
| NC score | 0.925002 (rank : 30) | θ value | 1.12514e-34 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
NSF_HUMAN
|
||||||
| NC score | 0.924363 (rank : 31) | θ value | 5.584e-34 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
KTNA1_HUMAN
|
||||||
| NC score | 0.923488 (rank : 32) | θ value | 1.51715e-39 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KTNA1_MOUSE
|
||||||
| NC score | 0.921292 (rank : 33) | θ value | 1.98146e-39 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
SPAST_HUMAN
|
||||||
| NC score | 0.917842 (rank : 34) | θ value | 7.29293e-34 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.916139 (rank : 35) | θ value | 2.12192e-33 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.909976 (rank : 36) | θ value | 5.22648e-32 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.909757 (rank : 37) | θ value | 2.34606e-32 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.907779 (rank : 38) | θ value | 4.72714e-33 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.907760 (rank : 39) | θ value | 4.72714e-33 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.897974 (rank : 40) | θ value | 1.28137e-46 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.880839 (rank : 41) | θ value | 1.51363e-47 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.871587 (rank : 42) | θ value | 2.51237e-26 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.866006 (rank : 43) | θ value | 7.30988e-26 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
BCS1_HUMAN
|
||||||
| NC score | 0.723579 (rank : 44) | θ value | 3.18553e-13 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| NC score | 0.715340 (rank : 45) | θ value | 4.16044e-13 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
TRP13_MOUSE
|
||||||
| NC score | 0.713040 (rank : 46) | θ value | 8.11959e-09 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| NC score | 0.709378 (rank : 47) | θ value | 1.80886e-08 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
ATD3A_HUMAN
|
||||||
| NC score | 0.630170 (rank : 48) | θ value | 1.53129e-07 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.620855 (rank : 49) | θ value | 1.38499e-08 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.612743 (rank : 50) | θ value | 5.26297e-08 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.444497 (rank : 51) | θ value | 0.000786445 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| NC score | 0.442680 (rank : 52) | θ value | 0.000786445 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.435324 (rank : 53) | θ value | 0.0193708 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
RUVB1_HUMAN
|
||||||
| NC score | 0.318976 (rank : 54) | θ value | 0.0330416 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| NC score | 0.318937 (rank : 55) | θ value | 0.0330416 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.262689 (rank : 56) | θ value | 0.00665767 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.258302 (rank : 57) | θ value | 0.0193708 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
RUVB2_HUMAN
|
||||||
| NC score | 0.122403 (rank : 58) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| NC score | 0.119852 (rank : 59) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.114273 (rank : 60) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
THNSL_MOUSE
|
||||||
| NC score | 0.106712 (rank : 61) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
THNSL_HUMAN
|
||||||
| NC score | 0.104392 (rank : 62) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.101199 (rank : 63) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RFC4_HUMAN
|
||||||
| NC score | 0.101004 (rank : 64) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.093553 (rank : 65) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC4_MOUSE
|
||||||
| NC score | 0.089972 (rank : 66) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
RFC5_MOUSE
|
||||||
| NC score | 0.079422 (rank : 67) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_HUMAN
|
||||||
| NC score | 0.078006 (rank : 68) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
KAD6_MOUSE
|
||||||
| NC score | 0.071031 (rank : 69) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.038758 (rank : 70) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
KAD2_HUMAN
|
||||||
| NC score | 0.037280 (rank : 71) | θ value | 0.163984 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54819, Q16856, Q5TIF7, Q8TCY2, Q8TCY3 | Gene names | AK2, ADK2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 2, mitochondrial (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
RENT1_MOUSE
|
||||||
| NC score | 0.026915 (rank : 72) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
RENT1_HUMAN
|
||||||
| NC score | 0.025380 (rank : 73) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
CKAP5_HUMAN
|
||||||
| NC score | 0.023277 (rank : 74) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14008, Q14668 | Gene names | CKAP5, KIAA0097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 5 (Colonic and hepatic tumor over- expressed protein) (Ch-TOG protein). | |||||
|
ICA69_HUMAN
|
||||||
| NC score | 0.022340 (rank : 75) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q05084, P78506, Q13824 | Gene names | ICA1 | |||
|
Domain Architecture |

|
|||||
| Description | Islet cell autoantigen 1 (69 kDa islet cell autoantigen) (ICA69) (p69) (Islet cell autoantigen p69) (ICAp69). | |||||
|
KAD2_MOUSE
|
||||||
| NC score | 0.018633 (rank : 76) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTP6, Q3TKI6, Q9CY37 | Gene names | Ak2, Ak-2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 2, mitochondrial (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
LC7L2_HUMAN
|
||||||
| NC score | 0.013786 (rank : 77) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
LC7L2_MOUSE
|
||||||
| NC score | 0.013591 (rank : 78) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
IF140_HUMAN
|
||||||
| NC score | 0.012195 (rank : 79) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RY7, O60332 | Gene names | IFT140, KIAA0590, WDTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 140 homolog (WD and tetratricopeptide repeats protein 2). | |||||
|
CDC27_HUMAN
|
||||||
| NC score | 0.011458 (rank : 80) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
NCOA7_HUMAN
|
||||||
| NC score | 0.010810 (rank : 81) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NI08, Q3LID6, Q4G0V1, Q5TF95, Q6IPQ4, Q6NE83, Q86T89, Q8N1W4 | Gene names | NCOA7, ERAP140, ESNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7 (140 kDa estrogen receptor-associated protein) (Estrogen nuclear receptor coactivator 1). | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.010465 (rank : 82) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
PALM_MOUSE
|
||||||
| NC score | 0.009454 (rank : 83) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.001860 (rank : 84) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.001797 (rank : 85) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
P55G_HUMAN
|
||||||
| NC score | 0.000590 (rank : 86) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | -0.001374 (rank : 87) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||