Please be patient as the page loads
|
WRIP1_HUMAN
|
||||||
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
WRIP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993788 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
RFC2_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 3) | NC score | 0.375213 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35250, P32846, Q9BU93 | Gene names | RFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RFC2_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 4) | NC score | 0.375113 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WUK4 | Gene names | Rfc2 | |||
|
Domain Architecture |
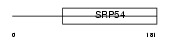
|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RFC5_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 5) | NC score | 0.363093 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 6) | NC score | 0.355493 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RAD18_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 7) | NC score | 0.139674 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NS91, Q58F55, Q9NRT6 | Gene names | RAD18, RNF73 | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (hRAD18) (hHR18) (RING finger protein 73). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 8) | NC score | 0.263251 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS6B_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 9) | NC score | 0.263845 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 10) | NC score | 0.264360 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
ATAD1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 11) | NC score | 0.263682 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 12) | NC score | 0.263682 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
PRS4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.258302 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 14) | NC score | 0.258302 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.253479 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
RFC4_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.289182 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
NVL_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.257470 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
RAD17_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.207250 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6NXW6, O88934, O89024 | Gene names | Rad17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17. | |||||
|
RFC4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.285609 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 20) | NC score | 0.050887 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
RAD18_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.114937 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
PRS6A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.249347 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.249560 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
RAD17_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.191162 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75943, O75714, Q7Z3S4, Q9UNK7, Q9UNR7, Q9UNR8, Q9UPF5 | Gene names | RAD17, R24L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17 (hRad17) (RF-C/activator 1 homolog). | |||||
|
TERA_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.247877 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.247872 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PRS10_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.249212 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.249212 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.099551 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.099554 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.048407 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
SPAST_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.232000 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPG7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.229795 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.231533 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.030909 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.230936 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.230921 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
DMWD_MOUSE
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.050159 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08274 | Gene names | Dmwd, Dm9 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophia myotonica WD repeat-containing protein (Dystrophia myotonica-containing WD repeat motif protein) (DMR-N9 protein). | |||||
|
KKCC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.009892 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5S9, Q9BQH3 | Gene names | CAMKK1, CAMKKA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 1 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase alpha) (CaM-kinase kinase alpha) (CaM-KK alpha) (CaMKK alpha) (CaMKK 1) (CaM-kinase IV kinase). | |||||
|
KTNA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.230493 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KTNA1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.230738 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
PRS7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.240116 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.240114 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.230381 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.230132 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
KATL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.228698 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.028670 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RFC3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.206210 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R323 | Gene names | Rfc3 | |||
|
Domain Architecture |
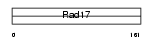
|
|||||
| Description | Replication factor C subunit 3 (Replication factor C 38 kDa subunit) (RFC38) (Activator 1 38 kDa subunit) (A1 38 kDa subunit) (RF-C 38 kDa subunit). | |||||
|
KKCC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.008663 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VBY2, Q9R054 | Gene names | Camkk1, Camkk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 1 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase alpha) (CaM-kinase kinase alpha) (CaM-KK alpha) (CaMKK alpha) (CaMKK 1) (CaM-kinase IV kinase). | |||||
|
RXINP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.057251 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RL1, Q5XKQ1, Q7Z3W7, Q8N5B9, Q9BZR1, Q9BZR5, Q9UHX7 | Gene names | RXRIP110, RAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110 (Receptor-associated protein 80) (Nuclear zinc finger protein RAP80). | |||||
|
ZAR1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.074882 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
KATL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.226333 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.229362 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
ZN174_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.003985 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15697, Q9BQ34 | Gene names | ZNF174, ZSCAN8 | |||
|
Domain Architecture |
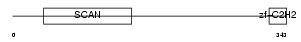
|
|||||
| Description | Zinc finger protein 174 (AW-1) (Zinc finger and SCAN domain-containing protein 8). | |||||
|
AFG32_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.217280 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.217113 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
NSF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.224279 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
NSF_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.224127 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
PRS8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.232252 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.232252 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.132789 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.024439 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.010451 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
SC6A5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.008087 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
BC11A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.010014 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H165, Q86W14, Q8WU92, Q96JL6, Q9H163, Q9H164, Q9H3G9, Q9NWA7 | Gene names | BCL11A, CTIP1, EVI9, KIAA1809 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11A (B-cell CLL/lymphoma 11A) (COUP-TF- interacting protein 1) (Ecotropic viral integration site 9 protein) (EVI-9). | |||||
|
BC11A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.012620 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYE3, Q80T89, Q8BLC7, Q8BLR4, Q8BWX3, Q921V4, Q9D0V2, Q9JIT4, Q9JLK8, Q9JLK9 | Gene names | Bcl11a, Ctip1, Evi9, Kiaa1809 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11A (B-cell CLL/lymphoma 11A) (COUP-TF- interacting protein 1) (Ecotropic viral integration site 9 protein) (EVI-9). | |||||
|
COA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.026483 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
KAD6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.075072 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.043683 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
AFG31_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.214964 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
BCS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.189770 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.184395 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.011834 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
KAD6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.076819 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
LYL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.014872 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
PEX5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.017512 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.220826 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
RFC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.187420 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P40938, O15252 | Gene names | RFC3 | |||
|
Domain Architecture |
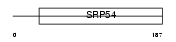
|
|||||
| Description | Replication factor C subunit 3 (Replication factor C 38 kDa subunit) (RFC38) (Activator 1 38 kDa subunit) (A1 38 kDa subunit) (RF-C 38 kDa subunit). | |||||
|
ZF276_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.003385 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.038760 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
KAD4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.027004 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR9, Q9R1X7 | Gene names | Ak3l1, Ak-4, Ak3b, Ak4 | |||
|
Domain Architecture |
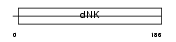
|
|||||
| Description | Adenylate kinase isoenzyme 4, mitochondrial (EC 2.7.4.3) (Adenylate kinase 3-like 1) (ATP-AMP transphosphorylase). | |||||
|
LONM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.136904 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.002183 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PZP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.016882 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20742, Q15273 | Gene names | PZP | |||
|
Domain Architecture |

|
|||||
| Description | Pregnancy zone protein precursor. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.020559 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SHC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.015743 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98083, Q3U2Q7, Q8BFY3, Q8K4C6, Q8K4C7 | Gene names | Shc1, Shc, ShcA | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 1 (SH2 domain protein C1) (Src homology 2 domain-containing-transforming protein C1). | |||||
|
SP6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.003943 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESX2, Q6VM22 | Gene names | Sp6, Epfn, Klf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14) (Epiprofin). | |||||
|
K2027_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.016010 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.006960 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
THNSL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.049431 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
ASPX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.017833 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.201705 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.205797 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.124542 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.123294 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
JERKY_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.012561 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75564, O75565 | Gene names | JRK, JH8 | |||
|
Domain Architecture |
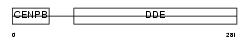
|
|||||
| Description | Jerky protein homolog. | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.023533 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SUSD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.008223 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.021421 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
TRPM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.006960 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.133821 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
ATD3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.137708 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.137180 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
FIGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.191123 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.192172 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
RFC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.120376 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RUVB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.078922 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.078912 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
TRP13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.161605 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
TRP13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.162263 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
YMEL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.202868 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.204001 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.993788 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
RFC2_HUMAN
|
||||||
| NC score | 0.375213 (rank : 3) | θ value | 1.99992e-07 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35250, P32846, Q9BU93 | Gene names | RFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RFC2_MOUSE
|
||||||
| NC score | 0.375113 (rank : 4) | θ value | 2.61198e-07 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WUK4 | Gene names | Rfc2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RFC5_HUMAN
|
||||||
| NC score | 0.363093 (rank : 5) | θ value | 3.77169e-06 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| NC score | 0.355493 (rank : 6) | θ value | 1.87187e-05 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC4_MOUSE
|
||||||
| NC score | 0.289182 (rank : 7) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
RFC4_HUMAN
|
||||||
| NC score | 0.285609 (rank : 8) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
PRS6B_MOUSE
|
||||||
| NC score | 0.264360 (rank : 9) | θ value | 0.00869519 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
PRS6B_HUMAN
|
||||||
| NC score | 0.263845 (rank : 10) | θ value | 0.00869519 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
ATAD1_HUMAN
|
||||||
| NC score | 0.263682 (rank : 11) | θ value | 0.0113563 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| NC score | 0.263682 (rank : 12) | θ value | 0.0113563 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.263251 (rank : 13) | θ value | 0.00665767 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS4_HUMAN
|
||||||
| NC score | 0.258302 (rank : 14) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| NC score | 0.258302 (rank : 15) | θ value | 0.0193708 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.257470 (rank : 16) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.253479 (rank : 17) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
PRS6A_MOUSE
|
||||||
| NC score | 0.249560 (rank : 18) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6A_HUMAN
|
||||||
| NC score | 0.249347 (rank : 19) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS10_HUMAN
|
||||||
| NC score | 0.249212 (rank : 20) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| NC score | 0.249212 (rank : 21) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
TERA_HUMAN
|
||||||
| NC score | 0.247877 (rank : 22) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| NC score | 0.247872 (rank : 23) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PRS7_HUMAN
|
||||||
| NC score | 0.240116 (rank : 24) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| NC score | 0.240114 (rank : 25) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS8_HUMAN
|
||||||
| NC score | 0.232252 (rank : 26) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| NC score | 0.232252 (rank : 27) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
SPAST_HUMAN
|
||||||
| NC score | 0.232000 (rank : 28) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.231533 (rank : 29) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.230936 (rank : 30) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.230921 (rank : 31) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
KTNA1_MOUSE
|
||||||
| NC score | 0.230738 (rank : 32) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
KTNA1_HUMAN
|
||||||
| NC score | 0.230493 (rank : 33) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.230381 (rank : 34) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.230132 (rank : 35) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
SPG7_HUMAN
|
||||||
| NC score | 0.229795 (rank : 36) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.229362 (rank : 37) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
KATL1_MOUSE
|
||||||
| NC score | 0.228698 (rank : 38) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_HUMAN
|
||||||
| NC score | 0.226333 (rank : 39) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
NSF_HUMAN
|
||||||
| NC score | 0.224279 (rank : 40) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
NSF_MOUSE
|
||||||
| NC score | 0.224127 (rank : 41) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.220826 (rank : 42) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
AFG32_HUMAN
|
||||||
| NC score | 0.217280 (rank : 43) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| NC score | 0.217113 (rank : 44) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
AFG31_MOUSE
|
||||||
| NC score | 0.214964 (rank : 45) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
RAD17_MOUSE
|
||||||
| NC score | 0.207250 (rank : 46) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6NXW6, O88934, O89024 | Gene names | Rad17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17. | |||||
|
RFC3_MOUSE
|
||||||
| NC score | 0.206210 (rank : 47) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R323 | Gene names | Rfc3 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 3 (Replication factor C 38 kDa subunit) (RFC38) (Activator 1 38 kDa subunit) (A1 38 kDa subunit) (RF-C 38 kDa subunit). | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.205797 (rank : 48) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.204001 (rank : 49) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
YMEL1_HUMAN
|
||||||
| NC score | 0.202868 (rank : 50) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.201705 (rank : 51) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.192172 (rank : 52) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
RAD17_HUMAN
|
||||||
| NC score | 0.191162 (rank : 53) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75943, O75714, Q7Z3S4, Q9UNK7, Q9UNR7, Q9UNR8, Q9UPF5 | Gene names | RAD17, R24L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17 (hRad17) (RF-C/activator 1 homolog). | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.191123 (rank : 54) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
BCS1_HUMAN
|
||||||
| NC score | 0.189770 (rank : 55) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
RFC3_HUMAN
|
||||||
| NC score | 0.187420 (rank : 56) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P40938, O15252 | Gene names | RFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 3 (Replication factor C 38 kDa subunit) (RFC38) (Activator 1 38 kDa subunit) (A1 38 kDa subunit) (RF-C 38 kDa subunit). | |||||
|
BCS1_MOUSE
|
||||||
| NC score | 0.184395 (rank : 57) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
TRP13_MOUSE
|
||||||
| NC score | 0.162263 (rank : 58) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| NC score | 0.161605 (rank : 59) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
RAD18_HUMAN
|
||||||
| NC score | 0.139674 (rank : 60) | θ value | 0.00390308 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NS91, Q58F55, Q9NRT6 | Gene names | RAD18, RNF73 | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (hRAD18) (hHR18) (RING finger protein 73). | |||||
|
ATD3A_HUMAN
|
||||||
| NC score | 0.137708 (rank : 61) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.137180 (rank : 62) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.136904 (rank : 63) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.133821 (rank : 64) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.132789 (rank : 65) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.124542 (rank : 66) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| NC score | 0.123294 (rank : 67) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.120376 (rank : 68) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RAD18_MOUSE
|
||||||
| NC score | 0.114937 (rank : 69) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.099554 (rank : 70) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.099551 (rank : 71) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
RUVB1_HUMAN
|
||||||
| NC score | 0.078922 (rank : 72) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| NC score | 0.078912 (rank : 73) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.076819 (rank : 74) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
KAD6_MOUSE
|
||||||
| NC score | 0.075072 (rank : 75) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
ZAR1_HUMAN
|
||||||
| NC score | 0.074882 (rank : 76) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
RXINP_HUMAN
|
||||||
| NC score | 0.057251 (rank : 77) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RL1, Q5XKQ1, Q7Z3W7, Q8N5B9, Q9BZR1, Q9BZR5, Q9UHX7 | Gene names | RXRIP110, RAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110 (Receptor-associated protein 80) (Nuclear zinc finger protein RAP80). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.050887 (rank : 78) | θ value | 0.0961366 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
DMWD_MOUSE
|
||||||
| NC score | 0.050159 (rank : 79) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08274 | Gene names | Dmwd, Dm9 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophia myotonica WD repeat-containing protein (Dystrophia myotonica-containing WD repeat motif protein) (DMR-N9 protein). | |||||
|
THNSL_HUMAN
|
||||||
| NC score | 0.049431 (rank : 80) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.048407 (rank : 81) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.043683 (rank : 82) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.038760 (rank : 83) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.030909 (rank : 84) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.028670 (rank : 85) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
KAD4_MOUSE
|
||||||
| NC score | 0.027004 (rank : 86) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR9, Q9R1X7 | Gene names | Ak3l1, Ak-4, Ak3b, Ak4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 4, mitochondrial (EC 2.7.4.3) (Adenylate kinase 3-like 1) (ATP-AMP transphosphorylase). | |||||
|
COA2_HUMAN
|
||||||
| NC score | 0.026483 (rank : 87) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.024439 (rank : 88) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.023533 (rank : 89) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.021421 (rank : 90) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.020559 (rank : 91) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ASPX_HUMAN
|
||||||
| NC score | 0.017833 (rank : 92) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
PEX5_MOUSE
|
||||||
| NC score | 0.017512 (rank : 93) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
PZP_HUMAN
|
||||||
| NC score | 0.016882 (rank : 94) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20742, Q15273 | Gene names | PZP | |||
|
Domain Architecture |

|
|||||
| Description | Pregnancy zone protein precursor. | |||||
|
K2027_HUMAN
|
||||||
| NC score | 0.016010 (rank : 95) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
SHC1_MOUSE
|
||||||
| NC score | 0.015743 (rank : 96) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98083, Q3U2Q7, Q8BFY3, Q8K4C6, Q8K4C7 | Gene names | Shc1, Shc, ShcA | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 1 (SH2 domain protein C1) (Src homology 2 domain-containing-transforming protein C1). | |||||
|
LYL1_MOUSE
|
||||||
| NC score | 0.014872 (rank : 97) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
BC11A_MOUSE
|
||||||
| NC score | 0.012620 (rank : 98) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYE3, Q80T89, Q8BLC7, Q8BLR4, Q8BWX3, Q921V4, Q9D0V2, Q9JIT4, Q9JLK8, Q9JLK9 | Gene names | Bcl11a, Ctip1, Evi9, Kiaa1809 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11A (B-cell CLL/lymphoma 11A) (COUP-TF- interacting protein 1) (Ecotropic viral integration site 9 protein) (EVI-9). | |||||
|
JERKY_HUMAN
|
||||||
| NC score | 0.012561 (rank : 99) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75564, O75565 | Gene names | JRK, JH8 | |||
|
Domain Architecture |

|
|||||
| Description | Jerky protein homolog. | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.011834 (rank : 100) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.010451 (rank : 101) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
BC11A_HUMAN
|
||||||
| NC score | 0.010014 (rank : 102) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H165, Q86W14, Q8WU92, Q96JL6, Q9H163, Q9H164, Q9H3G9, Q9NWA7 | Gene names | BCL11A, CTIP1, EVI9, KIAA1809 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11A (B-cell CLL/lymphoma 11A) (COUP-TF- interacting protein 1) (Ecotropic viral integration site 9 protein) (EVI-9). | |||||
|
KKCC1_HUMAN
|
||||||
| NC score | 0.009892 (rank : 103) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5S9, Q9BQH3 | Gene names | CAMKK1, CAMKKA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 1 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase alpha) (CaM-kinase kinase alpha) (CaM-KK alpha) (CaMKK alpha) (CaMKK 1) (CaM-kinase IV kinase). | |||||
|
KKCC1_MOUSE
|
||||||
| NC score | 0.008663 (rank : 104) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VBY2, Q9R054 | Gene names | Camkk1, Camkk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 1 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase alpha) (CaM-kinase kinase alpha) (CaM-KK alpha) (CaMKK alpha) (CaMKK 1) (CaM-kinase IV kinase). | |||||
|
SUSD4_MOUSE
|
||||||
| NC score | 0.008223 (rank : 105) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
SC6A5_MOUSE
|
||||||
| NC score | 0.008087 (rank : 106) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
TRPM2_HUMAN
|
||||||
| NC score | 0.006960 (rank : 107) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.006960 (rank : 108) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
ZN174_HUMAN
|
||||||
| NC score | 0.003985 (rank : 109) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15697, Q9BQ34 | Gene names | ZNF174, ZSCAN8 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 174 (AW-1) (Zinc finger and SCAN domain-containing protein 8). | |||||
|
SP6_MOUSE
|
||||||
| NC score | 0.003943 (rank : 110) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESX2, Q6VM22 | Gene names | Sp6, Epfn, Klf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14) (Epiprofin). | |||||
|
ZF276_HUMAN
|
||||||
| NC score | 0.003385 (rank : 111) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.002183 (rank : 112) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||