Please be patient as the page loads
|
RXINP_HUMAN
|
||||||
| SwissProt Accessions | Q96RL1, Q5XKQ1, Q7Z3W7, Q8N5B9, Q9BZR1, Q9BZR5, Q9UHX7 | Gene names | RXRIP110, RAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110 (Receptor-associated protein 80) (Nuclear zinc finger protein RAP80). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RXINP_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q96RL1, Q5XKQ1, Q7Z3W7, Q8N5B9, Q9BZR1, Q9BZR5, Q9UHX7 | Gene names | RXRIP110, RAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110 (Receptor-associated protein 80) (Nuclear zinc finger protein RAP80). | |||||
|
RXINP_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.956617 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5U5Q9, Q60811 | Gene names | Rxrip110, Rip110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110. | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 3) | NC score | 0.018609 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 0.125558 (rank : 4) | NC score | 0.072648 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
SHOC2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.025975 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ13, O76063 | Gene names | SHOC2, KIAA0862 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 0.163984 (rank : 6) | NC score | 0.059587 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
ENDD1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.112813 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C522, Q4VA96, Q8C5M1, Q8VE35 | Gene names | Endod1, Kiaa0830 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease domain-containing 1 protein precursor (EC 3.1.30.-). | |||||
|
ENDD1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.136776 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94919, Q8TAQ8 | Gene names | ENDOD1, KIAA0830 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease domain-containing 1 protein precursor (EC 3.1.30.-). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.061071 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.063359 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.037757 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.058360 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.057296 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.058218 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.038069 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.051408 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
TPX2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.093828 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULW0, Q9H1R4, Q9NRA3, Q9UFN9, Q9UL00, Q9Y2M1 | Gene names | TPX2, C20orf1, C20orf2, DIL2, HCA519 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Targeting protein for Xklp2 (Restricted expression proliferation- associated protein 100) (p100) (Differentially expressed in cancerous and noncancerous lung cells 2) (DIL-2) (Protein FLS353) (Hepatocellular carcinoma-associated antigen 519). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.039621 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
WNK3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.013867 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
SHOC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.022473 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88520, Q3UJH6, Q8BVL0 | Gene names | Shoc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
APC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.040989 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
CING_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.042595 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.040839 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
WRIP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.057251 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.056517 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.039074 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.046254 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.038188 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CN045_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.058229 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8ND07, Q9H5P8 | Gene names | C14orf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf45. | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.014917 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
PEDF_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.012014 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97298, O70629, O88691 | Gene names | Serpinf1, Pedf, Sdf3 | |||
|
Domain Architecture |
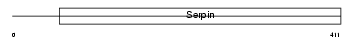
|
|||||
| Description | Pigment epithelium-derived factor precursor (PEDF) (Serpin-F1) (Stromal cell-derived factor 3) (SDF-3) (Caspin). | |||||
|
ZSWM2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.028606 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.012056 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
FBXW7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.009943 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
GLI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.004840 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||
|
GP158_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.024971 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.052507 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.013963 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
SFRS8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.037795 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
ZFP28_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.003514 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHY6, Q9BY30, Q9P2B6 | Gene names | ZFP28, KIAA1431 | |||
|
Domain Architecture |
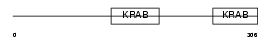
|
|||||
| Description | Zinc finger protein 28 homolog (Zfp-28) (Krueppel-like zinc finger factor X6). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.034838 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CING_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.041613 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.020700 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
K0555_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.041887 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
M3K4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.008877 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.047516 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MTMR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.013607 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
SKIL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.038679 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
TF2AA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.041730 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52655 | Gene names | GTF2A1, TF2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) (TFIIA-42) (TFIIAL) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
TF2AA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.040798 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PM3, Q8C812 | Gene names | Gtf2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.034063 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CDR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.037513 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01850, Q13977 | Gene names | CDR2, PCD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2 (Paraneoplastic cerebellar degeneration-associated antigen) (Major Yo paraneoplastic antigen). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.038668 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
LACTB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.030890 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P83111, P83096 | Gene names | LACTB | |||
|
Domain Architecture |
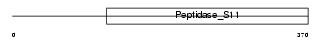
|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.037368 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.018841 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SPT16_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.036551 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y5B9, Q6GMT8, Q6P2F1, Q6PJM1, Q9NRX0 | Gene names | SUPT16H, FACT140, FACTP140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (hSPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin-specific transcription elongation factor 140 kDa subunit). | |||||
|
SPT16_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.036531 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q920B9, Q3TZ48, Q3UFH0, Q921H4 | Gene names | Supt16h, Fact140, Factp140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin- specific transcription elongation factor 140 kDa subunit). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.019948 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
ANR50_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.004698 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.035997 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
DESP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.029399 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.019301 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
ARP8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.023253 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H981, Q9H663 | Gene names | ACTR8 | |||
|
Domain Architecture |

|
|||||
| Description | Actin-related protein 8. | |||||
|
ARP8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.023339 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2S9, Q9CYC4 | Gene names | Actr8 | |||
|
Domain Architecture |

|
|||||
| Description | Actin-related protein 8. | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.011974 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.032592 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
HS90A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.023118 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.049003 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.024925 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
MAGB4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.006552 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |
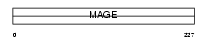
|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.048193 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MED12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.023880 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.021118 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
UBP53_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.013185 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.010796 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
CF152_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.030457 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
DCR1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.012981 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
HS90B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.019831 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
HS90B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.019063 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
MMTA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.018722 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BU76, Q6P5Y0, Q6ZTZ6, Q6ZWA6, Q8IZH3 | Gene names | MMTAG2, C1orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 (hMMTAG2). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.021602 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
PRDM4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.000931 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V63 | Gene names | Prdm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
RAD18_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.026304 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
RYBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.017861 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N488, Q9P2W5, Q9UMW4 | Gene names | RYBP, DEDAF, YEAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING1 and YY1-binding protein (Death effector domain-associated factor) (DED-associated factor) (YY1 and E4TF1-associated factor 1) (Apoptin-associating protein 1) (APAP-1). | |||||
|
WDHD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.009849 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
ZN462_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.002832 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JM2, Q8N408 | Gene names | ZNF462, KIAA1803 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 462. | |||||
|
RXINP_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q96RL1, Q5XKQ1, Q7Z3W7, Q8N5B9, Q9BZR1, Q9BZR5, Q9UHX7 | Gene names | RXRIP110, RAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110 (Receptor-associated protein 80) (Nuclear zinc finger protein RAP80). | |||||
|
RXINP_MOUSE
|
||||||
| NC score | 0.956617 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5U5Q9, Q60811 | Gene names | Rxrip110, Rip110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110. | |||||
|
ENDD1_HUMAN
|
||||||
| NC score | 0.136776 (rank : 3) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94919, Q8TAQ8 | Gene names | ENDOD1, KIAA0830 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease domain-containing 1 protein precursor (EC 3.1.30.-). | |||||
|
ENDD1_MOUSE
|
||||||
| NC score | 0.112813 (rank : 4) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C522, Q4VA96, Q8C5M1, Q8VE35 | Gene names | Endod1, Kiaa0830 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease domain-containing 1 protein precursor (EC 3.1.30.-). | |||||
|
TPX2_HUMAN
|
||||||
| NC score | 0.093828 (rank : 5) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULW0, Q9H1R4, Q9NRA3, Q9UFN9, Q9UL00, Q9Y2M1 | Gene names | TPX2, C20orf1, C20orf2, DIL2, HCA519 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Targeting protein for Xklp2 (Restricted expression proliferation- associated protein 100) (p100) (Differentially expressed in cancerous and noncancerous lung cells 2) (DIL-2) (Protein FLS353) (Hepatocellular carcinoma-associated antigen 519). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.072648 (rank : 6) | θ value | 0.125558 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.063359 (rank : 7) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.061071 (rank : 8) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.059587 (rank : 9) | θ value | 0.163984 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.058360 (rank : 10) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CN045_HUMAN
|
||||||
| NC score | 0.058229 (rank : 11) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8ND07, Q9H5P8 | Gene names | C14orf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf45. | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.058218 (rank : 12) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.057296 (rank : 13) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.057251 (rank : 14) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.056517 (rank : 15) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.052507 (rank : 16) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.051408 (rank : 17) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.049003 (rank : 18) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.048193 (rank : 19) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.047516 (rank : 20) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.046254 (rank : 21) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.042595 (rank : 22) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.041887 (rank : 23) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
TF2AA_HUMAN
|
||||||
| NC score | 0.041730 (rank : 24) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52655 | Gene names | GTF2A1, TF2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) (TFIIA-42) (TFIIAL) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.041613 (rank : 25) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.040989 (rank : 26) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.040839 (rank : 27) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
TF2AA_MOUSE
|
||||||
| NC score | 0.040798 (rank : 28) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PM3, Q8C812 | Gene names | Gtf2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.039621 (rank : 29) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.039074 (rank : 30) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SKIL_MOUSE
|
||||||
| NC score | 0.038679 (rank : 31) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.038668 (rank : 32) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.038188 (rank : 33) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.038069 (rank : 34) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
SFRS8_HUMAN
|
||||||
| NC score | 0.037795 (rank : 35) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.037757 (rank : 36) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
CDR2_HUMAN
|
||||||
| NC score | 0.037513 (rank : 37) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01850, Q13977 | Gene names | CDR2, PCD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2 (Paraneoplastic cerebellar degeneration-associated antigen) (Major Yo paraneoplastic antigen). | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.037368 (rank : 38) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
SPT16_HUMAN
|
||||||
| NC score | 0.036551 (rank : 39) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y5B9, Q6GMT8, Q6P2F1, Q6PJM1, Q9NRX0 | Gene names | SUPT16H, FACT140, FACTP140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (hSPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin-specific transcription elongation factor 140 kDa subunit). | |||||
|
SPT16_MOUSE
|
||||||
| NC score | 0.036531 (rank : 40) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q920B9, Q3TZ48, Q3UFH0, Q921H4 | Gene names | Supt16h, Fact140, Factp140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin- specific transcription elongation factor 140 kDa subunit). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.035997 (rank : 41) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.034838 (rank : 42) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.034063 (rank : 43) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.032592 (rank : 44) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
LACTB_HUMAN
|
||||||
| NC score | 0.030890 (rank : 45) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P83111, P83096 | Gene names | LACTB | |||
|
Domain Architecture |

|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.030457 (rank : 46) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.029399 (rank : 47) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
ZSWM2_MOUSE
|
||||||
| NC score | 0.028606 (rank : 48) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
RAD18_MOUSE
|
||||||
| NC score | 0.026304 (rank : 49) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
SHOC2_HUMAN
|
||||||
| NC score | 0.025975 (rank : 50) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ13, O76063 | Gene names | SHOC2, KIAA0862 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
GP158_MOUSE
|
||||||
| NC score | 0.024971 (rank : 51) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.024925 (rank : 52) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
MED12_HUMAN
|
||||||
| NC score | 0.023880 (rank : 53) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
ARP8_MOUSE
|
||||||
| NC score | 0.023339 (rank : 54) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2S9, Q9CYC4 | Gene names | Actr8 | |||
|
Domain Architecture |

|
|||||
| Description | Actin-related protein 8. | |||||
|
ARP8_HUMAN
|
||||||
| NC score | 0.023253 (rank : 55) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H981, Q9H663 | Gene names | ACTR8 | |||
|
Domain Architecture |

|
|||||
| Description | Actin-related protein 8. | |||||
|
HS90A_MOUSE
|
||||||
| NC score | 0.023118 (rank : 56) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
SHOC2_MOUSE
|
||||||
| NC score | 0.022473 (rank : 57) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88520, Q3UJH6, Q8BVL0 | Gene names | Shoc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.021602 (rank : 58) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.021118 (rank : 59) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.020700 (rank : 60) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.019948 (rank : 61) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
HS90B_HUMAN
|
||||||
| NC score | 0.019831 (rank : 62) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.019301 (rank : 63) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
HS90B_MOUSE
|
||||||
| NC score | 0.019063 (rank : 64) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.018841 (rank : 65) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
MMTA2_HUMAN
|
||||||
| NC score | 0.018722 (rank : 66) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BU76, Q6P5Y0, Q6ZTZ6, Q6ZWA6, Q8IZH3 | Gene names | MMTAG2, C1orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 (hMMTAG2). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.018609 (rank : 67) | θ value | 0.0113563 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
RYBP_HUMAN
|
||||||
| NC score | 0.017861 (rank : 68) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N488, Q9P2W5, Q9UMW4 | Gene names | RYBP, DEDAF, YEAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING1 and YY1-binding protein (Death effector domain-associated factor) (DED-associated factor) (YY1 and E4TF1-associated factor 1) (Apoptin-associating protein 1) (APAP-1). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.014917 (rank : 69) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.013963 (rank : 70) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
WNK3_HUMAN
|
||||||
| NC score | 0.013867 (rank : 71) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
MTMR3_HUMAN
|
||||||
| NC score | 0.013607 (rank : 72) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
UBP53_MOUSE
|
||||||
| NC score | 0.013185 (rank : 73) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
DCR1A_MOUSE
|
||||||
| NC score | 0.012981 (rank : 74) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.012056 (rank : 75) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
PEDF_MOUSE
|
||||||
| NC score | 0.012014 (rank : 76) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97298, O70629, O88691 | Gene names | Serpinf1, Pedf, Sdf3 | |||
|
Domain Architecture |

|
|||||
| Description | Pigment epithelium-derived factor precursor (PEDF) (Serpin-F1) (Stromal cell-derived factor 3) (SDF-3) (Caspin). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.011974 (rank : 77) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.010796 (rank : 78) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
FBXW7_HUMAN
|
||||||
| NC score | 0.009943 (rank : 79) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
WDHD1_HUMAN
|
||||||
| NC score | 0.009849 (rank : 80) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
M3K4_MOUSE
|
||||||
| NC score | 0.008877 (rank : 81) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
MAGB4_HUMAN
|
||||||
| NC score | 0.006552 (rank : 82) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
GLI1_MOUSE
|
||||||
| NC score | 0.004840 (rank : 83) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||
|
ANR50_HUMAN
|
||||||
| NC score | 0.004698 (rank : 84) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
ZFP28_HUMAN
|
||||||
| NC score | 0.003514 (rank : 85) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHY6, Q9BY30, Q9P2B6 | Gene names | ZFP28, KIAA1431 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 28 homolog (Zfp-28) (Krueppel-like zinc finger factor X6). | |||||
|
ZN462_HUMAN
|
||||||
| NC score | 0.002832 (rank : 86) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JM2, Q8N408 | Gene names | ZNF462, KIAA1803 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 462. | |||||
|
PRDM4_MOUSE
|
||||||
| NC score | 0.000931 (rank : 87) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V63 | Gene names | Prdm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||