Please be patient as the page loads
|
HS90A_MOUSE
|
||||||
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HS90A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987110 (rank : 2) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
HS90A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 155 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
HS90B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.979917 (rank : 3) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
HS90B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.979765 (rank : 4) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
ENPL_HUMAN
|
||||||
| θ value | 1.75921e-104 (rank : 5) | NC score | 0.922992 (rank : 6) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14625, Q96A97 | Gene names | HSP90B1, TRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (gp96 homolog) (Tumor rejection antigen 1). | |||||
|
ENPL_MOUSE
|
||||||
| θ value | 1.48926e-103 (rank : 6) | NC score | 0.923573 (rank : 5) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P08113, P11427 | Gene names | Hsp90b1, Tra-1, Tra1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (ERP99) (Polymorphic tumor rejection antigen 1) (Tumor rejection antigen gp96). | |||||
|
TRAP1_MOUSE
|
||||||
| θ value | 8.57503e-51 (rank : 7) | NC score | 0.846786 (rank : 8) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQN1, Q542I4 | Gene names | Trap1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
TRAP1_HUMAN
|
||||||
| θ value | 7.25919e-50 (rank : 8) | NC score | 0.848342 (rank : 7) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12931, O43642, O75235, Q9UHL5 | Gene names | TRAP1, HSP75 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 9) | NC score | 0.086891 (rank : 13) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 10) | NC score | 0.086547 (rank : 14) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.041686 (rank : 59) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 12) | NC score | 0.039763 (rank : 64) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
HSF2B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.153706 (rank : 9) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75031 | Gene names | HSF2BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock factor 2-binding protein. | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.052561 (rank : 32) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.065955 (rank : 16) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NIN_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.047542 (rank : 44) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.036755 (rank : 78) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.111579 (rank : 10) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 19) | NC score | 0.106115 (rank : 11) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TR150_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.064071 (rank : 18) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.061945 (rank : 20) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TR150_HUMAN
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.067124 (rank : 15) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.049655 (rank : 41) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.042665 (rank : 56) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
ANLN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.055492 (rank : 24) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
F10A1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.040818 (rank : 60) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.065045 (rank : 17) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.059933 (rank : 22) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.011362 (rank : 149) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.043706 (rank : 52) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.055104 (rank : 25) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
PRI1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.088097 (rank : 12) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20664 | Gene names | Prim1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA primase small subunit (EC 2.7.7.-) (DNA primase 49 kDa subunit) (p49). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.037592 (rank : 75) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.043858 (rank : 51) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
OXSR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.010755 (rank : 151) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95747, Q3LR53, Q7Z501, Q9UPQ1 | Gene names | OXSR1, KIAA1101, OSR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase OSR1 (EC 2.7.11.1) (Oxidative stress- responsive 1 protein). | |||||
|
OXSR1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.012006 (rank : 148) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9R2, Q8BVZ9, Q8C0B9 | Gene names | Oxsr1, Osr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase OSR1 (EC 2.7.11.1) (Oxidative stress- responsive 1 protein). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.040135 (rank : 62) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.034370 (rank : 91) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.021310 (rank : 123) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.043238 (rank : 54) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.048340 (rank : 42) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.048047 (rank : 43) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SURF2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.042318 (rank : 57) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09926 | Gene names | Surf2, Surf-2 | |||
|
Domain Architecture |
|
|||||
| Description | Surfeit locus protein 2 (Surf-2). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.033125 (rank : 97) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
F10A4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.038909 (rank : 67) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
F10A5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.038122 (rank : 71) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.045766 (rank : 49) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.050778 (rank : 39) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ST1E1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.024433 (rank : 117) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49888, Q8N6X5 | Gene names | SULT1E1, STE | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen sulfotransferase (EC 2.8.2.4) (Sulfotransferase, estrogen- preferring) (EST-1). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.037566 (rank : 76) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.047354 (rank : 45) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.026311 (rank : 114) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
S12A6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.018456 (rank : 133) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.018386 (rank : 134) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.045877 (rank : 48) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.051510 (rank : 35) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
ZN639_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.001165 (rank : 165) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UID6 | Gene names | ZNF639, ZASC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 639 (Zinc finger protein ZASC1) (Zinc finger protein ANC_2H01). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.038993 (rank : 66) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
GCP5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.024363 (rank : 118) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RT8, Q96PY8 | Gene names | TUBGCP5, GCP5, KIAA1899 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 5 (GCP-5). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.036468 (rank : 81) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HDGF_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.029767 (rank : 108) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.062308 (rank : 19) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.029274 (rank : 110) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.015358 (rank : 143) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.014779 (rank : 145) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.036162 (rank : 83) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.038016 (rank : 72) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
FGD4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.008450 (rank : 158) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.033374 (rank : 96) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SDPR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.046100 (rank : 47) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.052389 (rank : 33) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
STK39_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.007422 (rank : 161) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |
|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.043566 (rank : 53) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.034884 (rank : 88) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CCD55_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.037777 (rank : 74) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.037928 (rank : 73) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.036425 (rank : 82) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
HS105_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.018501 (rank : 132) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
IF2M_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.015935 (rank : 141) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |
|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.039330 (rank : 65) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
MPIP3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.031674 (rank : 103) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30307, Q96PL3, Q9H168, Q9H2E8, Q9H2E9, Q9H2F1 | Gene names | CDC25C | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 3 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25C). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.031164 (rank : 104) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NUDC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.038384 (rank : 69) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35685, Q3UJS7 | Gene names | Nudc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear migration protein nudC (Nuclear distribution protein C homolog) (Silica-induced gene 92 protein) (SIG-92). | |||||
|
PITX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.009802 (rank : 153) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99697, O60578, O60579, O60580, Q9BY17 | Gene names | PITX2, ARP1, RGS, RIEG, RIEG1 | |||
|
Domain Architecture |
|
|||||
| Description | Pituitary homeobox 2 (RIEG bicoid-related homeobox transcription factor) (Solurshin) (ALL1-responsive protein ARP1). | |||||
|
RBBP5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.016662 (rank : 139) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |
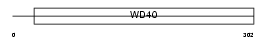
|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.042166 (rank : 58) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.054161 (rank : 26) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.033459 (rank : 95) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.027497 (rank : 113) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.050481 (rank : 40) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ACHA9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.007671 (rank : 160) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGM1, Q4W5A2, Q9NYV2 | Gene names | CHRNA9, NACHRA9 | |||
|
Domain Architecture |
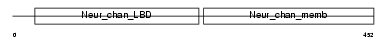
|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-9 precursor (Nicotinic acetylcholine receptor subunit alpha 9) (NACHR alpha 9). | |||||
|
ADA30_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.015462 (rank : 142) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.037563 (rank : 77) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.035986 (rank : 84) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.016392 (rank : 140) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CUL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.018826 (rank : 130) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.018824 (rank : 131) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
DGCR8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.020517 (rank : 126) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.025304 (rank : 115) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.036709 (rank : 79) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.034944 (rank : 87) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.030151 (rank : 106) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.022427 (rank : 122) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.045264 (rank : 50) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.040751 (rank : 61) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.020768 (rank : 125) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.035434 (rank : 85) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
AXN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.011328 (rank : 150) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
DGCR8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.019523 (rank : 129) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.039909 (rank : 63) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.036519 (rank : 80) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.028119 (rank : 112) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.031819 (rank : 101) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NUDC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.032437 (rank : 98) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y266, Q5QP31, Q5QP35, Q9H0N2, Q9Y2B6 | Gene names | NUDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear migration protein nudC (Nuclear distribution protein C homolog). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.018270 (rank : 136) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.014537 (rank : 146) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.034749 (rank : 89) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.030195 (rank : 105) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
AT10A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.015308 (rank : 144) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
CALU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.009240 (rank : 157) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
CALU_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.009438 (rank : 154) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35887 | Gene names | Calu | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.023339 (rank : 120) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.038202 (rank : 70) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.008134 (rank : 159) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CING_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.029874 (rank : 107) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CP135_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.034615 (rank : 90) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
LACTB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.017742 (rank : 137) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P83111, P83096 | Gene names | LACTB | |||
|
Domain Architecture |
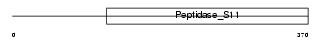
|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
NAF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.034332 (rank : 92) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
NGAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.019825 (rank : 128) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.032082 (rank : 100) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
PITX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.006607 (rank : 163) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97474, O08646, O70336, P97933, Q9JLA0, Q9QXB8, Q9R1V9, Q9Z141 | Gene names | Pitx2, Arp1, Brx1, Otlx2, Ptx2, Rgs | |||
|
Domain Architecture |
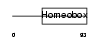
|
|||||
| Description | Pituitary homeobox 2 (Orthodenticle-like homeobox 2) (Solurshin) (ALL1-responsive protein ARP1) (BRX1 homeoprotein) (Paired-like homeodomain transcription factor Munc 30). | |||||
|
PMS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.024235 (rank : 119) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
RUFY2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.028986 (rank : 111) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
RXINP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.023118 (rank : 121) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RL1, Q5XKQ1, Q7Z3W7, Q8N5B9, Q9BZR1, Q9BZR5, Q9UHX7 | Gene names | RXRIP110, RAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110 (Receptor-associated protein 80) (Nuclear zinc finger protein RAP80). | |||||
|
STK39_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.006674 (rank : 162) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UEW8, O14774, Q9UER4 | Gene names | STK39, SPAK | |||
|
Domain Architecture |
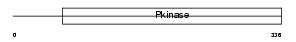
|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39) (DCHT). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.035085 (rank : 86) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
XLKD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.010215 (rank : 152) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5Y7, Q8TC18, Q9UNF4 | Gene names | XLKD1, CRSBP1, HAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Hyaluronic acid receptor) (Extracellular link domain-containing protein 1). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.047249 (rank : 46) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ZBT40_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | -0.000716 (rank : 166) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.042933 (rank : 55) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.031811 (rank : 102) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.032153 (rank : 99) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.034223 (rank : 93) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
DIAP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.009311 (rank : 155) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NSV4, Q5T2S7, Q5XKF6, Q6MZF0, Q6NUP0, Q86VS4, Q8NAV4 | Gene names | DIAPH3, DIAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3). | |||||
|
DYH9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.018345 (rank : 135) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
E2F6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.014400 (rank : 147) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54917 | Gene names | E2f6 | |||
|
Domain Architecture |
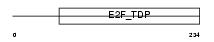
|
|||||
| Description | Transcription factor E2F6 (E2F-6) (E2F-binding site-modulating activity protein) (EMA). | |||||
|
FIBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.020067 (rank : 127) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI19 | Gene names | Fibp | |||
|
Domain Architecture |
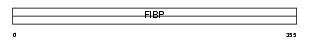
|
|||||
| Description | Acidic fibroblast growth factor intracellular-binding protein (aFGF intracellular-binding protein) (FGF-1 intracellular-binding protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.038623 (rank : 68) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.021221 (rank : 124) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
KLC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.009287 (rank : 156) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.034207 (rank : 94) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
NOL10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.025134 (rank : 116) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BSC4, Q53RC9, Q96TA5, Q9H7Y7, Q9H855 | Gene names | NOL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
SIL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.017698 (rank : 138) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H173, Q8N2L3 | Gene names | SIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleotide exchange factor SIL1 precursor (BiP-associated protein) (BAP). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.029299 (rank : 109) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ZA2G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.006227 (rank : 164) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64726 | Gene names | Azgp1 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc-alpha-2-glycoprotein precursor (Zn-alpha-2-glycoprotein) (Zn- alpha-2-GP). | |||||
|
CND3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.052249 (rank : 34) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.057671 (rank : 23) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.051142 (rank : 37) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.051310 (rank : 36) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.061277 (rank : 21) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.053858 (rank : 28) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.052981 (rank : 29) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
RBM25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.052622 (rank : 30) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.052568 (rank : 31) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.054063 (rank : 27) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.050856 (rank : 38) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
HS90A_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 155 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
HS90A_HUMAN
|
||||||
| NC score | 0.987110 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
HS90B_HUMAN
|
||||||
| NC score | 0.979917 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
HS90B_MOUSE
|
||||||
| NC score | 0.979765 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
ENPL_MOUSE
|
||||||
| NC score | 0.923573 (rank : 5) | θ value | 1.48926e-103 (rank : 6) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P08113, P11427 | Gene names | Hsp90b1, Tra-1, Tra1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (ERP99) (Polymorphic tumor rejection antigen 1) (Tumor rejection antigen gp96). | |||||
|
ENPL_HUMAN
|
||||||
| NC score | 0.922992 (rank : 6) | θ value | 1.75921e-104 (rank : 5) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14625, Q96A97 | Gene names | HSP90B1, TRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (gp96 homolog) (Tumor rejection antigen 1). | |||||
|
TRAP1_HUMAN
|
||||||
| NC score | 0.848342 (rank : 7) | θ value | 7.25919e-50 (rank : 8) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12931, O43642, O75235, Q9UHL5 | Gene names | TRAP1, HSP75 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
TRAP1_MOUSE
|
||||||
| NC score | 0.846786 (rank : 8) | θ value | 8.57503e-51 (rank : 7) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQN1, Q542I4 | Gene names | Trap1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
HSF2B_HUMAN
|
||||||
| NC score | 0.153706 (rank : 9) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75031 | Gene names | HSF2BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock factor 2-binding protein. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.111579 (rank : 10) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.106115 (rank : 11) | θ value | 0.0736092 (rank : 19) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PRI1_MOUSE
|
||||||
| NC score | 0.088097 (rank : 12) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20664 | Gene names | Prim1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA primase small subunit (EC 2.7.7.-) (DNA primase 49 kDa subunit) (p49). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.086891 (rank : 13) | θ value | 0.00298849 (rank : 9) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.086547 (rank : 14) | θ value | 0.00298849 (rank : 10) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TR150_HUMAN
|
||||||
| NC score | 0.067124 (rank : 15) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.065955 (rank : 16) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.065045 (rank : 17) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.064071 (rank : 18) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.062308 (rank : 19) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.061945 (rank : 20) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.061277 (rank : 21) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.059933 (rank : 22) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.057671 (rank : 23) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
ANLN_MOUSE
|
||||||
| NC score | 0.055492 (rank : 24) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.055104 (rank : 25) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.054161 (rank : 26) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.054063 (rank : 27) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.053858 (rank : 28) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.052981 (rank : 29) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.052622 (rank : 30) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.052568 (rank : 31) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.052561 (rank : 32) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.052389 (rank : 33) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
CND3_HUMAN
|
||||||
| NC score | 0.052249 (rank : 34) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.051510 (rank : 35) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.051310 (rank : 36) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.051142 (rank : 37) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.050856 (rank : 38) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.050778 (rank : 39) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.050481 (rank : 40) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.049655 (rank : 41) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.048340 (rank : 42) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.048047 (rank : 43) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.047542 (rank : 44) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.047354 (rank : 45) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.047249 (rank : 46) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
SDPR_HUMAN
|
||||||
| NC score | 0.046100 (rank : 47) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.045877 (rank : 48) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.045766 (rank : 49) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.045264 (rank : 50) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.043858 (rank : 51) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.043706 (rank : 52) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.043566 (rank : 53) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.043238 (rank : 54) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.042933 (rank : 55) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.042665 (rank : 56) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
SURF2_MOUSE
|
||||||
| NC score | 0.042318 (rank : 57) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09926 | Gene names | Surf2, Surf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 2 (Surf-2). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.042166 (rank : 58) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.041686 (rank : 59) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.040818 (rank : 60) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.040751 (rank : 61) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.040135 (rank : 62) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.039909 (rank : 63) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.039763 (rank : 64) | θ value | 0.0431538 (rank : 12) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.039330 (rank : 65) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.038993 (rank : 66) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
F10A4_HUMAN
|
||||||
| NC score | 0.038909 (rank : 67) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.038623 (rank : 68) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
NUDC_MOUSE
|
||||||
| NC score | 0.038384 (rank : 69) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35685, Q3UJS7 | Gene names | Nudc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear migration protein nudC (Nuclear distribution protein C homolog) (Silica-induced gene 92 protein) (SIG-92). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.038202 (rank : 70) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
F10A5_HUMAN
|
||||||
| NC score | 0.038122 (rank : 71) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.038016 (rank : 72) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.037928 (rank : 73) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.037777 (rank : 74) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.037592 (rank : 75) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.037566 (rank : 76) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.037563 (rank : 77) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.036755 (rank : 78) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.036709 (rank : 79) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.036519 (rank : 80) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.036468 (rank : 81) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.036425 (rank : 82) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.036162 (rank : 83) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.035986 (rank : 84) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.035434 (rank : 85) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.035085 (rank : 86) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.034944 (rank : 87) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.034884 (rank : 88) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.034749 (rank : 89) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.034615 (rank : 90) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.034370 (rank : 91) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.034332 (rank : 92) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.034223 (rank : 93) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.034207 (rank : 94) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.033459 (rank : 95) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.033374 (rank : 96) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.033125 (rank : 97) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
NUDC_HUMAN
|
||||||
| NC score | 0.032437 (rank : 98) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y266, Q5QP31, Q5QP35, Q9H0N2, Q9Y2B6 | Gene names | NUDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear migration protein nudC (Nuclear distribution protein C homolog). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.032153 (rank : 99) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.032082 (rank : 100) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.031819 (rank : 101) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.031811 (rank : 102) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
MPIP3_HUMAN
|
||||||
| NC score | 0.031674 (rank : 103) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30307, Q96PL3, Q9H168, Q9H2E8, Q9H2E9, Q9H2F1 | Gene names | CDC25C | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 3 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25C). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.031164 (rank : 104) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.030195 (rank : 105) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.030151 (rank : 106) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.029874 (rank : 107) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
HDGF_MOUSE
|
||||||
| NC score | 0.029767 (rank : 108) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.029299 (rank : 109) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.029274 (rank : 110) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
RUFY2_MOUSE
|
||||||
| NC score | 0.028986 (rank : 111) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.028119 (rank : 112) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.027497 (rank : 113) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.026311 (rank : 114) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.025304 (rank : 115) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
NOL10_HUMAN
|
||||||
| NC score | 0.025134 (rank : 116) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BSC4, Q53RC9, Q96TA5, Q9H7Y7, Q9H855 | Gene names | NOL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
ST1E1_HUMAN
|
||||||
| NC score | 0.024433 (rank : 117) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49888, Q8N6X5 | Gene names | SULT1E1, STE | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen sulfotransferase (EC 2.8.2.4) (Sulfotransferase, estrogen- preferring) (EST-1). | |||||
|
GCP5_HUMAN
|
||||||
| NC score | 0.024363 (rank : 118) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RT8, Q96PY8 | Gene names | TUBGCP5, GCP5, KIAA1899 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 5 (GCP-5). | |||||
|
PMS2_HUMAN
|
||||||
| NC score | 0.024235 (rank : 119) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.023339 (rank : 120) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
RXINP_HUMAN
|
||||||
| NC score | 0.023118 (rank : 121) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RL1, Q5XKQ1, Q7Z3W7, Q8N5B9, Q9BZR1, Q9BZR5, Q9UHX7 | Gene names | RXRIP110, RAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110 (Receptor-associated protein 80) (Nuclear zinc finger protein RAP80). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.022427 (rank : 122) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.021310 (rank : 123) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.021221 (rank : 124) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.020768 (rank : 125) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
DGCR8_MOUSE
|
||||||
| NC score | 0.020517 (rank : 126) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
FIBP_MOUSE
|
||||||
| NC score | 0.020067 (rank : 127) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI19 | Gene names | Fibp | |||
|
Domain Architecture |

|
|||||
| Description | Acidic fibroblast growth factor intracellular-binding protein (aFGF intracellular-binding protein) (FGF-1 intracellular-binding protein). | |||||
|
NGAP_HUMAN
|
||||||
| NC score | 0.019825 (rank : 128) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
DGCR8_HUMAN
|
||||||
| NC score | 0.019523 (rank : 129) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
CUL1_HUMAN
|
||||||
| NC score | 0.018826 (rank : 130) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| NC score | 0.018824 (rank : 131) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
HS105_HUMAN
|
||||||
| NC score | 0.018501 (rank : 132) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
S12A6_HUMAN
|
||||||
| NC score | 0.018456 (rank : 133) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| NC score | 0.018386 (rank : 134) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
DYH9_HUMAN
|
||||||
| NC score | 0.018345 (rank : 135) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.018270 (rank : 136) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
LACTB_HUMAN
|
||||||
| NC score | 0.017742 (rank : 137) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P83111, P83096 | Gene names | LACTB | |||
|
Domain Architecture |

|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
SIL1_HUMAN
|
||||||
| NC score | 0.017698 (rank : 138) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H173, Q8N2L3 | Gene names | SIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleotide exchange factor SIL1 precursor (BiP-associated protein) (BAP). | |||||
|
RBBP5_HUMAN
|
||||||
| NC score | 0.016662 (rank : 139) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.016392 (rank : 140) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
IF2M_MOUSE
|
||||||
| NC score | 0.015935 (rank : 141) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
ADA30_HUMAN
|
||||||
| NC score | 0.015462 (rank : 142) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.015358 (rank : 143) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
AT10A_HUMAN
|
||||||
| NC score | 0.015308 (rank : 144) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.014779 (rank : 145) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.014537 (rank : 146) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
E2F6_MOUSE
|
||||||
| NC score | 0.014400 (rank : 147) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54917 | Gene names | E2f6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F6 (E2F-6) (E2F-binding site-modulating activity protein) (EMA). | |||||
|
OXSR1_MOUSE
|
||||||
| NC score | 0.012006 (rank : 148) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9R2, Q8BVZ9, Q8C0B9 | Gene names | Oxsr1, Osr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase OSR1 (EC 2.7.11.1) (Oxidative stress- responsive 1 protein). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.011362 (rank : 149) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
AXN2_MOUSE
|
||||||
| NC score | 0.011328 (rank : 150) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
OXSR1_HUMAN
|
||||||
| NC score | 0.010755 (rank : 151) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95747, Q3LR53, Q7Z501, Q9UPQ1 | Gene names | OXSR1, KIAA1101, OSR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase OSR1 (EC 2.7.11.1) (Oxidative stress- responsive 1 protein). | |||||
|
XLKD1_HUMAN
|
||||||
| NC score | 0.010215 (rank : 152) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5Y7, Q8TC18, Q9UNF4 | Gene names | XLKD1, CRSBP1, HAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Hyaluronic acid receptor) (Extracellular link domain-containing protein 1). | |||||
|
PITX2_HUMAN
|
||||||
| NC score | 0.009802 (rank : 153) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99697, O60578, O60579, O60580, Q9BY17 | Gene names | PITX2, ARP1, RGS, RIEG, RIEG1 | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary homeobox 2 (RIEG bicoid-related homeobox transcription factor) (Solurshin) (ALL1-responsive protein ARP1). | |||||
|
CALU_MOUSE
|
||||||
| NC score | 0.009438 (rank : 154) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35887 | Gene names | Calu | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin). | |||||
|
DIAP3_HUMAN
|
||||||
| NC score | 0.009311 (rank : 155) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NSV4, Q5T2S7, Q5XKF6, Q6MZF0, Q6NUP0, Q86VS4, Q8NAV4 | Gene names | DIAPH3, DIAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3). | |||||
|
KLC4_HUMAN
|
||||||
| NC score | 0.009287 (rank : 156) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
CALU_HUMAN
|
||||||
| NC score | 0.009240 (rank : 157) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
FGD4_HUMAN
|
||||||
| NC score | 0.008450 (rank : 158) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.008134 (rank : 159) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
ACHA9_HUMAN
|
||||||
| NC score | 0.007671 (rank : 160) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGM1, Q4W5A2, Q9NYV2 | Gene names | CHRNA9, NACHRA9 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-9 precursor (Nicotinic acetylcholine receptor subunit alpha 9) (NACHR alpha 9). | |||||
|
STK39_MOUSE
|
||||||
| NC score | 0.007422 (rank : 161) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |

|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
STK39_HUMAN
|
||||||
| NC score | 0.006674 (rank : 162) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UEW8, O14774, Q9UER4 | Gene names | STK39, SPAK | |||
|
Domain Architecture |

|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39) (DCHT). | |||||
|
PITX2_MOUSE
|
||||||
| NC score | 0.006607 (rank : 163) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97474, O08646, O70336, P97933, Q9JLA0, Q9QXB8, Q9R1V9, Q9Z141 | Gene names | Pitx2, Arp1, Brx1, Otlx2, Ptx2, Rgs | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary homeobox 2 (Orthodenticle-like homeobox 2) (Solurshin) (ALL1-responsive protein ARP1) (BRX1 homeoprotein) (Paired-like homeodomain transcription factor Munc 30). | |||||
|
ZA2G_MOUSE
|
||||||
| NC score | 0.006227 (rank : 164) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64726 | Gene names | Azgp1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-alpha-2-glycoprotein precursor (Zn-alpha-2-glycoprotein) (Zn- alpha-2-GP). | |||||
|
ZN639_HUMAN
|
||||||
| NC score | 0.001165 (rank : 165) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UID6 | Gene names | ZNF639, ZASC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 639 (Zinc finger protein ZASC1) (Zinc finger protein ANC_2H01). | |||||
|
ZBT40_HUMAN
|
||||||
| NC score | -0.000716 (rank : 166) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 155 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||