Please be patient as the page loads
|
MMTA2_HUMAN
|
||||||
| SwissProt Accessions | Q9BU76, Q6P5Y0, Q6ZTZ6, Q6ZWA6, Q8IZH3 | Gene names | MMTAG2, C1orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 (hMMTAG2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MMTA2_HUMAN
|
||||||
| θ value | 1.1019e-106 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BU76, Q6P5Y0, Q6ZTZ6, Q6ZWA6, Q8IZH3 | Gene names | MMTAG2, C1orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 (hMMTAG2). | |||||
|
MMTA2_MOUSE
|
||||||
| θ value | 4.22625e-74 (rank : 2) | NC score | 0.959643 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LX5, Q9CSK6 | Gene names | Mmtag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 homolog. | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 3) | NC score | 0.077730 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 4) | NC score | 0.041587 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.21417 (rank : 5) | NC score | 0.086139 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 6) | NC score | 0.076578 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 7) | NC score | 0.094012 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
ERCC6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.028528 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.018691 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.057190 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.023600 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.032397 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
NEIL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.046513 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4Q6, Q80V58, Q9CYT9 | Gene names | Neil1, Nei1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1). | |||||
|
CS029_HUMAN
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.036555 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUQ7, O75229, Q7LE08, Q9BTA6, Q9Y5A4 | Gene names | C19orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 (NY-REN-24 antigen). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.060317 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SEMG2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.042011 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.039055 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
RNC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.055186 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
WDR60_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.049270 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.015737 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
RRP5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.064535 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
MECP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.035079 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |
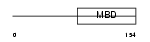
|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
TTBK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.015467 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.015800 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
CT151_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.037064 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |
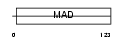
|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.006901 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
MCPH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.022907 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEM0, Q66GU1, Q9H9C7 | Gene names | MCPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.015249 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
ZN292_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.016155 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.010094 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.051364 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
IPP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.023410 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
JARD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.014347 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
LATS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.006174 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
LMO6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.007736 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |
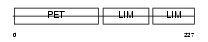
|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
LYAR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.036518 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
PTN6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.005076 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
RXINP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.018722 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RL1, Q5XKQ1, Q7Z3W7, Q8N5B9, Q9BZR1, Q9BZR5, Q9UHX7 | Gene names | RXRIP110, RAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110 (Receptor-associated protein 80) (Nuclear zinc finger protein RAP80). | |||||
|
TERF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.025003 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54274, Q15553, Q93029 | Gene names | TERF1, PIN2, TRF, TRF1 | |||
|
Domain Architecture |
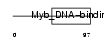
|
|||||
| Description | Telomeric repeat-binding factor 1 (TTAGGG repeat-binding factor 1) (NIMA-interacting protein 2) (Telomeric protein Pin2/TRF1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.031137 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.050369 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.051284 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
MMTA2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.1019e-106 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BU76, Q6P5Y0, Q6ZTZ6, Q6ZWA6, Q8IZH3 | Gene names | MMTAG2, C1orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 (hMMTAG2). | |||||
|
MMTA2_MOUSE
|
||||||
| NC score | 0.959643 (rank : 2) | θ value | 4.22625e-74 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LX5, Q9CSK6 | Gene names | Mmtag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 homolog. | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.094012 (rank : 3) | θ value | 0.279714 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.086139 (rank : 4) | θ value | 0.21417 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.077730 (rank : 5) | θ value | 0.0252991 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.076578 (rank : 6) | θ value | 0.279714 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
RRP5_HUMAN
|
||||||
| NC score | 0.064535 (rank : 7) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.060317 (rank : 8) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.057190 (rank : 9) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RNC_HUMAN
|
||||||
| NC score | 0.055186 (rank : 10) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.051364 (rank : 11) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.051284 (rank : 12) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.050369 (rank : 13) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
WDR60_HUMAN
|
||||||
| NC score | 0.049270 (rank : 14) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
NEIL1_MOUSE
|
||||||
| NC score | 0.046513 (rank : 15) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4Q6, Q80V58, Q9CYT9 | Gene names | Neil1, Nei1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1). | |||||
|
SEMG2_HUMAN
|
||||||
| NC score | 0.042011 (rank : 16) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.041587 (rank : 17) | θ value | 0.0961366 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.039055 (rank : 18) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
CT151_HUMAN
|
||||||
| NC score | 0.037064 (rank : 19) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
CS029_HUMAN
|
||||||
| NC score | 0.036555 (rank : 20) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUQ7, O75229, Q7LE08, Q9BTA6, Q9Y5A4 | Gene names | C19orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 (NY-REN-24 antigen). | |||||
|
LYAR_MOUSE
|
||||||
| NC score | 0.036518 (rank : 21) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
MECP2_HUMAN
|
||||||
| NC score | 0.035079 (rank : 22) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.032397 (rank : 23) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.031137 (rank : 24) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ERCC6_HUMAN
|
||||||
| NC score | 0.028528 (rank : 25) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
TERF1_HUMAN
|
||||||
| NC score | 0.025003 (rank : 26) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54274, Q15553, Q93029 | Gene names | TERF1, PIN2, TRF, TRF1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 1 (TTAGGG repeat-binding factor 1) (NIMA-interacting protein 2) (Telomeric protein Pin2/TRF1). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.023600 (rank : 27) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
IPP4_HUMAN
|
||||||
| NC score | 0.023410 (rank : 28) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
MCPH1_HUMAN
|
||||||
| NC score | 0.022907 (rank : 29) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEM0, Q66GU1, Q9H9C7 | Gene names | MCPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
RXINP_HUMAN
|
||||||
| NC score | 0.018722 (rank : 30) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RL1, Q5XKQ1, Q7Z3W7, Q8N5B9, Q9BZR1, Q9BZR5, Q9UHX7 | Gene names | RXRIP110, RAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110 (Receptor-associated protein 80) (Nuclear zinc finger protein RAP80). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.018691 (rank : 31) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ZN292_HUMAN
|
||||||
| NC score | 0.016155 (rank : 32) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | 0.015800 (rank : 33) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.015737 (rank : 34) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
TTBK2_HUMAN
|
||||||
| NC score | 0.015467 (rank : 35) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.015249 (rank : 36) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
JARD2_HUMAN
|
||||||
| NC score | 0.014347 (rank : 37) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.010094 (rank : 38) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
LMO6_HUMAN
|
||||||
| NC score | 0.007736 (rank : 39) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.006901 (rank : 40) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
LATS1_HUMAN
|
||||||
| NC score | 0.006174 (rank : 41) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
PTN6_HUMAN
|
||||||
| NC score | 0.005076 (rank : 42) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||