Please be patient as the page loads
|
LBXCO_MOUSE
|
||||||
| SwissProt Accessions | Q8BX46, Q3UYA4, Q5W8I2, Q8C0T2 | Gene names | Lbxcor1, Corl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1 (Transcriptional corepressor Corl1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LBXCO_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.965001 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
LBXCO_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q8BX46, Q3UYA4, Q5W8I2, Q8C0T2 | Gene names | Lbxcor1, Corl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1 (Transcriptional corepressor Corl1). | |||||
|
SKI_HUMAN
|
||||||
| θ value | 2.05525e-28 (rank : 3) | NC score | 0.594219 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12755, Q5SYT7 | Gene names | SKI | |||
|
Domain Architecture |
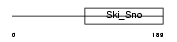
|
|||||
| Description | Ski oncogene (C-ski). | |||||
|
SKI_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 4) | NC score | 0.594113 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q60698, Q8VIL5 | Gene names | Ski | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ski oncogene (C-ski). | |||||
|
SKIL_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 5) | NC score | 0.496149 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
SKIL_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 6) | NC score | 0.546497 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12757, O00464, P12756, Q07501 | Gene names | SKIL, SNO | |||
|
Domain Architecture |
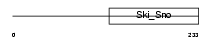
|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
DACH2_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 7) | NC score | 0.391458 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q925Q8, Q8BMA0 | Gene names | Dach2 | |||
|
Domain Architecture |
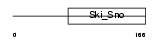
|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
DACH2_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 8) | NC score | 0.379392 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96NX9, Q8NAY3, Q8ND17, Q96N55 | Gene names | DACH2 | |||
|
Domain Architecture |
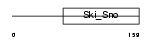
|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
DACH1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 9) | NC score | 0.346990 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
DACH1_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 10) | NC score | 0.344902 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 11) | NC score | 0.089032 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
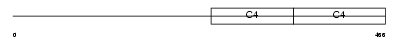
CO4A4_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 12) | NC score | 0.068250 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |
|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 13) | NC score | 0.056665 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.074201 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CP135_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.070674 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.049733 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.066600 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.027433 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.069350 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.038717 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
HIP1R_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.072094 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
SALL1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.010422 (rank : 164) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ER74, Q920R5 | Gene names | Sall1, Sal3 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein Spalt-3) (Sal-3) (MSal-3). | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.070105 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.059218 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.027060 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
AFF2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.050038 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.069482 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CP135_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.057605 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CO4A6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.062244 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14031, Q12823, Q14053, Q5JYH6, Q9NQM5, Q9NTX3, Q9UJ76, Q9UMG6, Q9Y4L4 | Gene names | COL4A6 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-6(IV) chain precursor. | |||||
|
CO9A2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.065172 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.079727 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.037474 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.028769 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.037137 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.055332 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.028638 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.073171 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.071726 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.056547 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TBCD4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.025564 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BYJ6, Q5DU23, Q66JU2, Q6P2M2, Q8BMH6, Q8BXM2 | Gene names | Tbc1d4, Kiaa0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.061070 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
DAAM2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.046209 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
DYHC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.056953 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.056884 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.031303 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LMNB2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.040435 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P21619, P48680, Q8CGB1 | Gene names | Lmnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
MSI2H_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.017704 (rank : 150) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96DH6, Q7Z6M7, Q8N9T4 | Gene names | MSI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
PDE4A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.023429 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
AATK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.007453 (rank : 167) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||
|
ASH2L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.031530 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.039465 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CING_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.059949 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.021821 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
MISS_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.038878 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.048955 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SH3K1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.032766 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96B97, Q8IWX6, Q8IX98, Q96RN4, Q9NYR0 | Gene names | SH3KBP1, CIN85 | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (Cbl-interacting protein of 85 kDa) (Human Src-family kinase-binding protein 1) (HSB-1) (CD2-binding protein 3) (CD2BP3). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.046158 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DAAM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.043139 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
EGR2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.004465 (rank : 173) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.005842 (rank : 168) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.080490 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
PDE4A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.021338 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PLCB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.035958 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.027446 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
CE290_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.049169 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CN145_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.057204 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
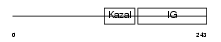
IBP7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.015674 (rank : 154) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
RAB3I_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.044622 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.022369 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
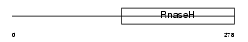
RNH1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.053656 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60930, O60523, O60857, Q57Z93, Q5U0C1, Q6FHD4 | Gene names | RNASEH1, RNH1 | |||
|
Domain Architecture |
|
|||||
| Description | Ribonuclease H1 (EC 3.1.26.4) (RNase H1) (Ribonuclease H type II). | |||||
|
S12A5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.016175 (rank : 152) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
WDHD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.025865 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.053262 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.052168 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MSI2H_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.014980 (rank : 155) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920Q6, Q8BQ90, Q920Q7 | Gene names | Msi2, Msi2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.051673 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.023561 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.039682 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.039632 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
STAM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.020365 (rank : 149) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
TMCC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.033381 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94876, Q68E06, Q8IXM8 | Gene names | TMCC1, KIAA0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
UN93B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.022216 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCW4, O89077, Q710D2, Q8CIK1, Q8R3D0 | Gene names | Unc93b1, Unc93b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC93 homolog B1 (UNC-93B protein). | |||||
|
CHIC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.037490 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CBW7, O08904, Q8K3S5 | Gene names | Chic1, Brx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 1 protein (Brain X-linked protein). | |||||
|
GATA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.012668 (rank : 159) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15976, Q96GB8 | Gene names | GATA1, ERYF1, GF1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.058034 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.014437 (rank : 157) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
LMNB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.039086 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.029133 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RAD9A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.027597 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0F6, Q8QZZ3 | Gene names | Rad9a, Rad9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint control protein RAD9A (EC 3.1.11.2) (DNA repair exonuclease rad9 homolog A) (Rad9-like protein) (mRAD9). | |||||
|
CING_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.051900 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CO5A3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.058278 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.043687 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
E2F2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.017576 (rank : 151) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14209 | Gene names | E2F2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.045481 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.054874 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
KCNK9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.010203 (rank : 165) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
PDZD8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.030118 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
PITM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.009856 (rank : 166) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.046598 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.024273 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
RUFY3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.054780 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D394, Q3U348, Q3UF96, Q6PE64, Q8VD10 | Gene names | Rufy3, D5Bwg0860e, Ripx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
SH3K1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.026382 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.020589 (rank : 148) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.010428 (rank : 163) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
WDHD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.021940 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
Z585B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | -0.000118 (rank : 176) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q52M93, Q8IZD3, Q96JW6 | Gene names | ZNF585B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 585B (zinc finger protein 41-like protein). | |||||
|
ZBTB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.004705 (rank : 171) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H5J0 | Gene names | ZBTB3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 3. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.021269 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
CAN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.005131 (rank : 170) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
DNJC9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.024092 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
E2F2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.015942 (rank : 153) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56931, Q8BID0 | Gene names | E2f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.020869 (rank : 147) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
JIP3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.046928 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
MEF2C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.013856 (rank : 158) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CFN5, Q8R0H1, Q9D7L0, Q9QW20 | Gene names | Mef2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myocyte-specific enhancer factor 2C. | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.005188 (rank : 169) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
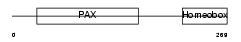
PAX7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.002329 (rank : 175) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23759 | Gene names | PAX7, HUP1 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-7 (HUP1). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.024697 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.023886 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.004666 (rank : 172) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
RUFY3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.053546 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7L099, O94948, Q9UI00 | Gene names | RUFY3, KIAA0871 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
TMCC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.025564 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q69ZZ6, Q8CEF4 | Gene names | Tmcc1, Kiaa0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.029259 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
CO2A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.054681 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |
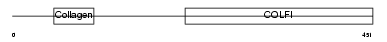
|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO2A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.056017 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
COBA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.054202 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.053775 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.012484 (rank : 160) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.040712 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
HEM6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.024970 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36551, Q14060, Q8IZ45, Q96AF3 | Gene names | CPOX, CPO, CPX | |||
|
Domain Architecture |

|
|||||
| Description | Coproporphyrinogen III oxidase, mitochondrial precursor (EC 1.3.3.3) (Coproporphyrinogenase) (Coprogen oxidase) (COX). | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.014787 (rank : 156) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
K0555_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.039359 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
NPHP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.034280 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.054199 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.064251 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.036338 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.003419 (rank : 174) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.054644 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TACC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.030928 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
TMC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.010830 (rank : 162) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
TRIM8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.011212 (rank : 161) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.050548 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.053992 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CN145_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.054448 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.050399 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO1A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.051806 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.051300 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.051300 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.057279 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
COGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.051595 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
COHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.055831 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
COHA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.051445 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
COIA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.050641 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
COJA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050613 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.064732 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.063922 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CTGE5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.058264 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
ELN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.058454 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
ELN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.055998 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54320 | Gene names | Eln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
HOOK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.058377 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
MACOI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.050590 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.052135 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.053059 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
NEMO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.060460 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.051861 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.055195 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.061555 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.050276 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
RABE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.053676 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.054667 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.056737 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.050376 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.050477 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.068636 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
TUFT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.061705 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
TUFT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.059668 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
ZN291_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.051687 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
LBXCO_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q8BX46, Q3UYA4, Q5W8I2, Q8C0T2 | Gene names | Lbxcor1, Corl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1 (Transcriptional corepressor Corl1). | |||||
|
LBXCO_HUMAN
|
||||||
| NC score | 0.965001 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
SKI_HUMAN
|
||||||
| NC score | 0.594219 (rank : 3) | θ value | 2.05525e-28 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12755, Q5SYT7 | Gene names | SKI | |||
|
Domain Architecture |

|
|||||
| Description | Ski oncogene (C-ski). | |||||
|
SKI_MOUSE
|
||||||
| NC score | 0.594113 (rank : 4) | θ value | 3.50572e-28 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q60698, Q8VIL5 | Gene names | Ski | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ski oncogene (C-ski). | |||||
|
SKIL_HUMAN
|
||||||
| NC score | 0.546497 (rank : 5) | θ value | 4.01107e-24 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12757, O00464, P12756, Q07501 | Gene names | SKIL, SNO | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
SKIL_MOUSE
|
||||||
| NC score | 0.496149 (rank : 6) | θ value | 3.07116e-24 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
DACH2_MOUSE
|
||||||
| NC score | 0.391458 (rank : 7) | θ value | 4.76016e-09 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q925Q8, Q8BMA0 | Gene names | Dach2 | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
DACH2_HUMAN
|
||||||
| NC score | 0.379392 (rank : 8) | θ value | 6.87365e-08 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96NX9, Q8NAY3, Q8ND17, Q96N55 | Gene names | DACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
DACH1_HUMAN
|
||||||
| NC score | 0.346990 (rank : 9) | θ value | 1.87187e-05 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
DACH1_MOUSE
|
||||||
| NC score | 0.344902 (rank : 10) | θ value | 5.44631e-05 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.089032 (rank : 11) | θ value | 0.000786445 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.080490 (rank : 12) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.079727 (rank : 13) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.074201 (rank : 14) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.073171 (rank : 15) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
HIP1R_HUMAN
|
||||||
| NC score | 0.072094 (rank : 16) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.071726 (rank : 17) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.070674 (rank : 18) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.070105 (rank : 19) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.069482 (rank : 20) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.069350 (rank : 21) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.068636 (rank : 22) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
CO4A4_HUMAN
|
||||||
| NC score | 0.068250 (rank : 23) | θ value | 0.00869519 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.066600 (rank : 24) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CO9A2_MOUSE
|
||||||
| NC score | 0.065172 (rank : 25) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.064732 (rank : 26) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.064251 (rank : 27) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.063922 (rank : 28) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CO4A6_HUMAN
|
||||||
| NC score | 0.062244 (rank : 29) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14031, Q12823, Q14053, Q5JYH6, Q9NQM5, Q9NTX3, Q9UJ76, Q9UMG6, Q9Y4L4 | Gene names | COL4A6 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-6(IV) chain precursor. | |||||
|
TUFT1_HUMAN
|
||||||
| NC score | 0.061705 (rank : 30) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.061555 (rank : 31) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.061070 (rank : 32) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
NEMO_HUMAN
|
||||||
| NC score | 0.060460 (rank : 33) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.059949 (rank : 34) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
TUFT1_MOUSE
|
||||||
| NC score | 0.059668 (rank : 35) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.059218 (rank : 36) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
ELN_HUMAN
|
||||||
| NC score | 0.058454 (rank : 37) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
HOOK2_HUMAN
|
||||||
| NC score | 0.058377 (rank : 38) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
CO5A3_HUMAN
|
||||||
| NC score | 0.058278 (rank : 39) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
CTGE5_HUMAN
|
||||||
| NC score | 0.058264 (rank : 40) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.058034 (rank : 41) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.057605 (rank : 42) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.057279 (rank : 43) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.057204 (rank : 44) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
DYHC_HUMAN
|
||||||
| NC score | 0.056953 (rank : 45) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| NC score | 0.056884 (rank : 46) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.056737 (rank : 47) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.056665 (rank : 48) | θ value | 0.00869519 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.056547 (rank : 49) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CO2A1_MOUSE
|
||||||
| NC score | 0.056017 (rank : 50) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
ELN_MOUSE
|
||||||
| NC score | 0.055998 (rank : 51) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54320 | Gene names | Eln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
COHA1_HUMAN
|
||||||
| NC score | 0.055831 (rank : 52) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.055332 (rank : 53) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.055195 (rank : 54) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.054874 (rank : 55) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
RUFY3_MOUSE
|
||||||
| NC score | 0.054780 (rank : 56) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D394, Q3U348, Q3UF96, Q6PE64, Q8VD10 | Gene names | Rufy3, D5Bwg0860e, Ripx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
CO2A1_HUMAN
|
||||||
| NC score | 0.054681 (rank : 57) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.054667 (rank : 58) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.054644 (rank : 59) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.054448 (rank : 60) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.054202 (rank : 61) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.054199 (rank : 62) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.053992 (rank : 63) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
COBA1_MOUSE
|
||||||
| NC score | 0.053775 (rank : 64) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.053676 (rank : 65) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RNH1_HUMAN
|
||||||
| NC score | 0.053656 (rank : 66) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60930, O60523, O60857, Q57Z93, Q5U0C1, Q6FHD4 | Gene names | RNASEH1, RNH1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease H1 (EC 3.1.26.4) (RNase H1) (Ribonuclease H type II). | |||||
|
RUFY3_HUMAN
|
||||||
| NC score | 0.053546 (rank : 67) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7L099, O94948, Q9UI00 | Gene names | RUFY3, KIAA0871 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.053262 (rank : 68) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.053059 (rank : 69) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.052168 (rank : 70) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.052135 (rank : 71) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.051900 (rank : 72) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.051861 (rank : 73) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CO1A1_MOUSE
|
||||||
| NC score | 0.051806 (rank : 74) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
ZN291_HUMAN
|
||||||
| NC score | 0.051687 (rank : 75) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.051673 (rank : 76) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
COGA1_HUMAN
|
||||||
| NC score | 0.051595 (rank : 77) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.051445 (rank : 78) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.051300 (rank : 79) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.051300 (rank : 80) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.050641 (rank : 81) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
COJA1_HUMAN
|
||||||
| NC score | 0.050613 (rank : 82) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
MACOI_HUMAN
|
||||||
| NC score | 0.050590 (rank : 83) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.050548 (rank : 84) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.050477 (rank : 85) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.050399 (rank : 86) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.050376 (rank : 87) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.050276 (rank : 88) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
AFF2_MOUSE
|
||||||
| NC score | 0.050038 (rank : 89) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.049733 (rank : 90) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.049169 (rank : 91) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.048955 (rank : 92) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
JIP3_MOUSE
|
||||||
| NC score | 0.046928 (rank : 93) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.046598 (rank : 94) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
DAAM2_MOUSE
|
||||||
| NC score | 0.046209 (rank : 95) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.046158 (rank : 96) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.045481 (rank : 97) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
RAB3I_HUMAN
|
||||||
| NC score | 0.044622 (rank : 98) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.043687 (rank : 99) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
DAAM2_HUMAN
|
||||||
| NC score | 0.043139 (rank : 100) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.040712 (rank : 101) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
LMNB2_MOUSE
|
||||||
| NC score | 0.040435 (rank : 102) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P21619, P48680, Q8CGB1 | Gene names | Lmnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.039682 (rank : 103) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.039632 (rank : 104) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.039465 (rank : 105) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.039359 (rank : 106) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
LMNB2_HUMAN
|
||||||
| NC score | 0.039086 (rank : 107) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.038878 (rank : 108) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.038717 (rank : 109) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CHIC1_MOUSE
|
||||||
| NC score | 0.037490 (rank : 110) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CBW7, O08904, Q8K3S5 | Gene names | Chic1, Brx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 1 protein (Brain X-linked protein). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.037474 (rank : 111) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.037137 (rank : 112) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.036338 (rank : 113) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
PLCB2_HUMAN
|
||||||
| NC score | 0.035958 (rank : 114) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
NPHP3_HUMAN
|
||||||
| NC score | 0.034280 (rank : 115) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
TMCC1_HUMAN
|
||||||
| NC score | 0.033381 (rank : 116) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94876, Q68E06, Q8IXM8 | Gene names | TMCC1, KIAA0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
SH3K1_HUMAN
|
||||||
| NC score | 0.032766 (rank : 117) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96B97, Q8IWX6, Q8IX98, Q96RN4, Q9NYR0 | Gene names | SH3KBP1, CIN85 | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (Cbl-interacting protein of 85 kDa) (Human Src-family kinase-binding protein 1) (HSB-1) (CD2-binding protein 3) (CD2BP3). | |||||
|
ASH2L_MOUSE
|
||||||
| NC score | 0.031530 (rank : 118) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.031303 (rank : 119) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TACC3_MOUSE
|
||||||
| NC score | 0.030928 (rank : 120) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
PDZD8_HUMAN
|
||||||
| NC score | 0.030118 (rank : 121) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.029259 (rank : 122) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.029133 (rank : 123) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.028769 (rank : 124) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.028638 (rank : 125) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
RAD9A_MOUSE
|
||||||
| NC score | 0.027597 (rank : 126) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0F6, Q8QZZ3 | Gene names | Rad9a, Rad9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint control protein RAD9A (EC 3.1.11.2) (DNA repair exonuclease rad9 homolog A) (Rad9-like protein) (mRAD9). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.027446 (rank : 127) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.027433 (rank : 128) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.027060 (rank : 129) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
SH3K1_MOUSE
|
||||||
| NC score | 0.026382 (rank : 130) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
WDHD1_MOUSE
|
||||||
| NC score | 0.025865 (rank : 131) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
TBCD4_MOUSE
|
||||||
| NC score | 0.025564 (rank : 132) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BYJ6, Q5DU23, Q66JU2, Q6P2M2, Q8BMH6, Q8BXM2 | Gene names | Tbc1d4, Kiaa0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
TMCC1_MOUSE
|
||||||
| NC score | 0.025564 (rank : 133) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q69ZZ6, Q8CEF4 | Gene names | Tmcc1, Kiaa0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
HEM6_HUMAN
|
||||||
| NC score | 0.024970 (rank : 134) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36551, Q14060, Q8IZ45, Q96AF3 | Gene names | CPOX, CPO, CPX | |||
|
Domain Architecture |

|
|||||
| Description | Coproporphyrinogen III oxidase, mitochondrial precursor (EC 1.3.3.3) (Coproporphyrinogenase) (Coprogen oxidase) (COX). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.024697 (rank : 135) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.024273 (rank : 136) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
DNJC9_HUMAN
|
||||||
| NC score | 0.024092 (rank : 137) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.023886 (rank : 138) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.023561 (rank : 139) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
PDE4A_MOUSE
|
||||||
| NC score | 0.023429 (rank : 140) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.022369 (rank : 141) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
UN93B_MOUSE
|
||||||
| NC score | 0.022216 (rank : 142) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCW4, O89077, Q710D2, Q8CIK1, Q8R3D0 | Gene names | Unc93b1, Unc93b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC93 homolog B1 (UNC-93B protein). | |||||
|
WDHD1_HUMAN
|
||||||
| NC score | 0.021940 (rank : 143) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.021821 (rank : 144) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
PDE4A_HUMAN
|
||||||
| NC score | 0.021338 (rank : 145) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.021269 (rank : 146) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.020869 (rank : 147) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.020589 (rank : 148) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
STAM2_MOUSE
|
||||||
| NC score | 0.020365 (rank : 149) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
MSI2H_HUMAN
|
||||||
| NC score | 0.017704 (rank : 150) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96DH6, Q7Z6M7, Q8N9T4 | Gene names | MSI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
E2F2_HUMAN
|
||||||
| NC score | 0.017576 (rank : 151) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14209 | Gene names | E2F2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
S12A5_MOUSE
|
||||||
| NC score | 0.016175 (rank : 152) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
E2F2_MOUSE
|
||||||
| NC score | 0.015942 (rank : 153) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56931, Q8BID0 | Gene names | E2f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
IBP7_HUMAN
|
||||||
| NC score | 0.015674 (rank : 154) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
MSI2H_MOUSE
|
||||||
| NC score | 0.014980 (rank : 155) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920Q6, Q8BQ90, Q920Q7 | Gene names | Msi2, Msi2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.014787 (rank : 156) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.014437 (rank : 157) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
MEF2C_MOUSE
|
||||||
| NC score | 0.013856 (rank : 158) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CFN5, Q8R0H1, Q9D7L0, Q9QW20 | Gene names | Mef2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myocyte-specific enhancer factor 2C. | |||||
|
GATA1_HUMAN
|
||||||
| NC score | 0.012668 (rank : 159) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15976, Q96GB8 | Gene names | GATA1, ERYF1, GF1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.012484 (rank : 160) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
TRIM8_HUMAN
|
||||||
| NC score | 0.011212 (rank : 161) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
TMC2_HUMAN
|
||||||
| NC score | 0.010830 (rank : 162) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.010428 (rank : 163) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
SALL1_MOUSE
|
||||||
| NC score | 0.010422 (rank : 164) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ER74, Q920R5 | Gene names | Sall1, Sal3 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein Spalt-3) (Sal-3) (MSal-3). | |||||
|
KCNK9_MOUSE
|
||||||
| NC score | 0.010203 (rank : 165) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
PITM1_MOUSE
|
||||||
| NC score | 0.009856 (rank : 166) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
AATK_MOUSE
|
||||||
| NC score | 0.007453 (rank : 167) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||
|
GLI2_HUMAN
|
||||||
| NC score | 0.005842 (rank : 168) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.005188 (rank : 169) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
CAN3_HUMAN
|
||||||
| NC score | 0.005131 (rank : 170) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
ZBTB3_HUMAN
|
||||||
| NC score | 0.004705 (rank : 171) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H5J0 | Gene names | ZBTB3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 3. | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.004666 (rank : 172) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
EGR2_HUMAN
|
||||||
| NC score | 0.004465 (rank : 173) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.003419 (rank : 174) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
PAX7_HUMAN
|
||||||
| NC score | 0.002329 (rank : 175) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23759 | Gene names | PAX7, HUP1 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-7 (HUP1). | |||||
|
Z585B_HUMAN
|
||||||
| NC score | -0.000118 (rank : 176) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q52M93, Q8IZD3, Q96JW6 | Gene names | ZNF585B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 585B (zinc finger protein 41-like protein). | |||||