Please be patient as the page loads
|
ZDHC5_MOUSE
|
||||||
| SwissProt Accessions | Q8VDZ4, Q69ZB5, Q8R2X7 | Gene names | Zdhhc5, Kiaa1748 | |||
|
Domain Architecture |
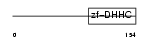
|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZDHC5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994149 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9C0B5, Q6ZMF0, Q8TAK8, Q9H923, Q9UFI7 | Gene names | ZDHHC5, KIAA1748, ZNF375 | |||
|
Domain Architecture |
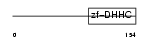
|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5) (Zinc finger protein 375). | |||||
|
ZDHC5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8VDZ4, Q69ZB5, Q8R2X7 | Gene names | Zdhhc5, Kiaa1748 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5). | |||||
|
ZDHC8_MOUSE
|
||||||
| θ value | 1.56551e-161 (rank : 3) | NC score | 0.961304 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
ZDHC8_HUMAN
|
||||||
| θ value | 5.03607e-160 (rank : 4) | NC score | 0.966014 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |
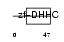
|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
ZDHC9_HUMAN
|
||||||
| θ value | 9.16162e-53 (rank : 5) | NC score | 0.924738 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y397, Q59EK4, Q5JSW5, Q8WWS7, Q9BPY4, Q9NSP0, Q9NVL0, Q9NVR6 | Gene names | ZDHHC9, ZNF379 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9) (Zinc finger protein 379). | |||||
|
ZDHC9_MOUSE
|
||||||
| θ value | 9.16162e-53 (rank : 6) | NC score | 0.924959 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59268, Q5BL19 | Gene names | Zdhhc9 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9). | |||||
|
ZDH14_HUMAN
|
||||||
| θ value | 3.98324e-48 (rank : 7) | NC score | 0.912224 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IZN3, Q5JS07, Q5JS08, Q6PHS4, Q8IZN2, Q9H7F1 | Gene names | ZDHHC14 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
ZDH14_MOUSE
|
||||||
| θ value | 3.98324e-48 (rank : 8) | NC score | 0.913243 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BQQ1, Q8BNR2, Q8CFN0 | Gene names | Zdhhc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
ZDH18_HUMAN
|
||||||
| θ value | 2.85459e-46 (rank : 9) | NC score | 0.923114 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NUE0, Q5JYH0, Q9H020 | Gene names | ZDHHC18 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC18 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 18) (DHHC-18). | |||||
|
ZDH19_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 10) | NC score | 0.909078 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVZ1 | Gene names | ZDHHC19 | |||
|
Domain Architecture |
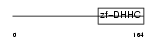
|
|||||
| Description | Probable palmitoyltransferase ZDHHC19 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 19) (DHHC-19). | |||||
|
ZDH19_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 11) | NC score | 0.895848 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q810M5 | Gene names | Zdhhc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC19 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 19) (DHHC-19). | |||||
|
ZDH20_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 12) | NC score | 0.805096 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5W0Z9, Q6NVU8 | Gene names | ZDHHC20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC20 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 20) (DHHC-20). | |||||
|
ZDH15_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 13) | NC score | 0.809290 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96MV8, Q3SY30, Q6UWH3 | Gene names | ZDHHC15 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
ZDH15_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 14) | NC score | 0.808407 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BGJ0, Q3TV63, Q8BMB7 | Gene names | Zdhhc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
ZDHC2_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 15) | NC score | 0.795196 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UIJ5 | Gene names | ZDHHC2, REAM, REC, ZNF372 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC2 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 2) (DHHC-2) (Zinc finger protein 372) (Reduced expression associated with metastasis protein) (Ream) (Reduced expression in cancer protein) (Rec). | |||||
|
ZDHC2_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 16) | NC score | 0.795974 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59267 | Gene names | Zdhhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC2 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 2) (DHHC-2). | |||||
|
ZDH20_MOUSE
|
||||||
| θ value | 3.51386e-20 (rank : 17) | NC score | 0.798687 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5Y5T1, Q3TDL1, Q8VCL6, Q9D3Q8 | Gene names | Zdhhc20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC20 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 20) (DHHC-20). | |||||
|
ZDH17_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 18) | NC score | 0.447774 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
ZDH17_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 19) | NC score | 0.453012 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |
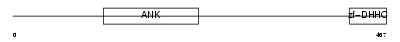
|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
ZDHC3_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 20) | NC score | 0.804842 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8R173, Q6EMK3 | Gene names | Zdhhc3, Godz, Gramp1 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC3 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 3) (DHHC-3) (Golgi-specific DHHC zinc finger protein) (GABA-A receptor-associated membrane protein 1). | |||||
|
ZDHC3_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 21) | NC score | 0.799430 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NYG2, Q53A17, Q96BL0 | Gene names | ZDHHC3, ZNF373 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC3 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 3) (DHHC-3) (Zinc finger protein 373) (DHHC1 protein). | |||||
|
ZDHC4_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 22) | NC score | 0.826766 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D6H5, Q8R5E0 | Gene names | Zdhhc4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC4 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 4) (DHHC-4). | |||||
|
ZDHC7_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 23) | NC score | 0.787458 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NXF8, Q8WV42, Q9NVD8 | Gene names | ZDHHC7, ZNF370 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC7 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 7) (DHHC-7) (Zinc finger protein 370). | |||||
|
ZDHC4_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 24) | NC score | 0.824590 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NPG8, Q53EV7, Q6FIB5, Q9H0R9 | Gene names | ZDHHC4, ZNF374 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC4 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 4) (DHHC-4) (Zinc finger protein 374). | |||||
|
ZDH16_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 25) | NC score | 0.805311 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ESG8, Q3TI22, Q3UA59, Q91XC5 | Gene names | Zdhhc16, Aph2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC16 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 16) (DHHC-16) (Ablphilin-2). | |||||
|
ZDHC7_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 26) | NC score | 0.787574 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91WU6, Q3TC41, Q6EMK2, Q8K1H6 | Gene names | Zdhhc7, Gramp2 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC7 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 7) (DHHC-7) (GABA-A receptor-associated membrane protein 2). | |||||
|
ZDHC6_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 27) | NC score | 0.791121 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H6R6, Q53G45, Q96IV7, Q9H605 | Gene names | ZDHHC6, ZNF376 | |||
|
Domain Architecture |
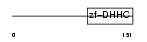
|
|||||
| Description | Probable palmitoyltransferase ZDHHC6 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 6) (DHHC-6) (Zinc finger protein 376) (Transmembrane protein H4). | |||||
|
ZDHC6_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 28) | NC score | 0.793103 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CPV7, Q3U3I8, Q3UT70, Q544H1 | Gene names | Zdhhc6 | |||
|
Domain Architecture |
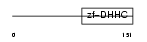
|
|||||
| Description | Probable palmitoyltransferase ZDHHC6 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 6) (DHHC-6) (H4 homolog). | |||||
|
ZDH12_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 29) | NC score | 0.850547 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VC90, Q9CZ24, Q9D0T6 | Gene names | Zdhhc12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC12 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 12) (DHHC-12). | |||||
|
ZDH12_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 30) | NC score | 0.842884 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96GR4, Q5T265, Q5T267, Q5T268, Q86VT5, Q96T09 | Gene names | ZDHHC12, ZNF400 | |||
|
Domain Architecture |
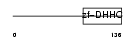
|
|||||
| Description | Probable palmitoyltransferase ZDHHC12 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 12) (DHHC-12) (Zinc finger protein 400). | |||||
|
ZDH16_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 31) | NC score | 0.795297 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q969W1, Q5JTG7, Q5JTH0, Q8N4Z6, Q8WY84, Q9BSV3 | Gene names | ZDHHC16 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC16 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 16) (DHHC-16). | |||||
|
ZDH11_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 32) | NC score | 0.785159 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H8X9 | Gene names | ZDHHC11, ZNF399 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC11 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 11) (DHHC-11) (Zinc finger protein 399). | |||||
|
ZDH23_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 33) | NC score | 0.768476 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5Y5T3, Q3UH65, Q3UVA2 | Gene names | Zdhhc23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC23 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 23) (DHHC-23) (DHHC-containing protein 11). | |||||
|
ZDH13_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 34) | NC score | 0.436033 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9CWU2, Q3UK32, Q3UKV1 | Gene names | Zdhhc13, Hip14l | |||
|
Domain Architecture |
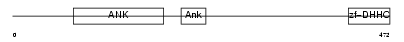
|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein). | |||||
|
ZDH21_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 35) | NC score | 0.792896 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IVQ6, Q5VWG1 | Gene names | ZDHHC21 | |||
|
Domain Architecture |
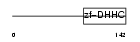
|
|||||
| Description | Probable palmitoyltransferase ZDHHC21 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 21) (DHHC-21). | |||||
|
ZDH21_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 36) | NC score | 0.788821 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D270, Q6EMK1, Q80XQ3 | Gene names | Zdhhc21, Gramp3 | |||
|
Domain Architecture |
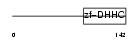
|
|||||
| Description | Probable palmitoyltransferase ZDHHC21 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 21) (DHHC-21) (GABA-A receptor-associated membrane protein 3). | |||||
|
ZDH13_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 37) | NC score | 0.430432 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8IUH4, Q7Z2D3, Q86VK2, Q9NV30, Q9NV99 | Gene names | ZDHHC13, HIP14L, HIP3RP | |||
|
Domain Architecture |
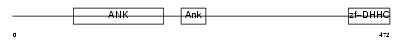
|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein) (Huntingtin- interacting protein HIP3RP) (Putative NF-kappa-B-activating protein 209) (Putative MAPK-activating protein PM03). | |||||
|
ZDHC1_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 38) | NC score | 0.737231 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WTX9, O15461 | Gene names | ZDHHC1, C16orf1, ZNF377 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC1 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 1) (DHHC-1) (Zinc finger protein 377) (DHHC- domain-containing cysteine-rich protein 1). | |||||
|
ZDH23_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 39) | NC score | 0.748681 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IYP9 | Gene names | ZDHHC23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC23 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 23) (DHHC-23). | |||||
|
ZDHC1_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 40) | NC score | 0.742022 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8R0N9, Q8BJ24 | Gene names | Zdhhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC1 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 1) (DHHC-1). | |||||
|
ZDH24_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 41) | NC score | 0.776996 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6UX98, Q6PEW7, Q9BSJ0 | Gene names | ZDHHC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC24 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 24) (DHHC-24). | |||||
|
ZDH24_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 42) | NC score | 0.769757 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6IR37, Q3TAY4, Q3TDF1, Q3TF09, Q9CS66 | Gene names | Zdhhc24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC24 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 24) (DHHC-24). | |||||
|
ZDH22_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 43) | NC score | 0.712032 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N966 | Gene names | ZDHHC22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative palmitoyltransferase ZDHHC22 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 22) (DHHC-22). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 44) | NC score | 0.040362 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 45) | NC score | 0.051376 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 46) | NC score | 0.050842 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 47) | NC score | 0.016481 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 48) | NC score | 0.027148 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
LPIN1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 49) | NC score | 0.029372 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZP3, Q9CQI2, Q9JLG6 | Gene names | Lpin1, Fld | |||
|
Domain Architecture |
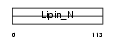
|
|||||
| Description | Lipin-1 (Fatty liver dystrophy protein). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 0.21417 (rank : 50) | NC score | 0.008550 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
IP3KB_HUMAN
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.027491 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.060124 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.045749 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
IL16_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.028910 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.004472 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.013267 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.030224 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.028253 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.024159 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.026075 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.015957 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
FYB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.034129 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
LPIN3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.020882 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |
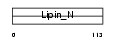
|
|||||
| Description | Lipin-3. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.030495 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.024454 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.016262 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
AKTS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.039312 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96B36, Q96BI4, Q96IK7, Q96NG2, Q9BWR5 | Gene names | AKT1S1, PRAS40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (40 kDa proline-rich AKT substrate). | |||||
|
AKTS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.042343 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D1F4 | Gene names | Akt1s1, Pras | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (Proline-rich AKT substrate). | |||||
|
AMOL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.006750 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
GAK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.027802 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.021315 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.027976 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.027931 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.028008 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.027943 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MLTK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | -0.002862 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.011523 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.026721 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.050986 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | -0.001441 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
O51D1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | -0.002150 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGF3 | Gene names | OR51D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 51D1 (Olfactory receptor OR11-14). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | -0.000136 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.026425 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.027595 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.027306 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
HME1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.001326 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09065 | Gene names | En1, En-1 | |||
|
Domain Architecture |
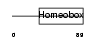
|
|||||
| Description | Homeobox protein engrailed-1 (Mo-En-1). | |||||
|
OCLN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.016560 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61146, Q544A7 | Gene names | Ocln, Ocl | |||
|
Domain Architecture |

|
|||||
| Description | Occludin. | |||||
|
SEM4A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.007094 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
PHLB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.018452 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.008097 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
GAK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.026617 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
K0562_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.015621 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80V31, Q6ZQ95 | Gene names | Kiaa0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.015737 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
PRPC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.045485 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
TTLL4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.007400 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
UBQL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.006724 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5U9 | Gene names | Ubqln3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.021876 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CLK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | -0.002613 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49761, Q9BRS3, Q9BUJ7 | Gene names | CLK3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase CLK3 (EC 2.7.12.1) (CDC-like kinase 3). | |||||
|
DUET_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | -0.003527 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.008635 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NOS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.003892 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0J4, Q64208 | Gene names | Nos1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.014624 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
THUM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.015421 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J36, Q3U6Y3 | Gene names | Thumpd1 | |||
|
Domain Architecture |

|
|||||
| Description | THUMP domain-containing protein 1. | |||||
|
CE005_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.002273 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.010456 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
GLI3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | -0.004607 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.013647 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.003921 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.008968 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.006374 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
SEM6C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.003151 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SLIK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.004821 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94933 | Gene names | SLITRK3, KIAA0848 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLIT and NTRK-like protein 3 precursor. | |||||
|
ZCHC4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.082485 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKW4, Q3UTX6, Q9D2I1 | Gene names | Zcchc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
PRPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.056666 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
ZCHC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.112223 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H5U6, Q5IW78, Q96AN7 | Gene names | ZCCHC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
ZDHC5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8VDZ4, Q69ZB5, Q8R2X7 | Gene names | Zdhhc5, Kiaa1748 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5). | |||||
|
ZDHC5_HUMAN
|
||||||
| NC score | 0.994149 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9C0B5, Q6ZMF0, Q8TAK8, Q9H923, Q9UFI7 | Gene names | ZDHHC5, KIAA1748, ZNF375 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5) (Zinc finger protein 375). | |||||
|
ZDHC8_HUMAN
|
||||||
| NC score | 0.966014 (rank : 3) | θ value | 5.03607e-160 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
ZDHC8_MOUSE
|
||||||
| NC score | 0.961304 (rank : 4) | θ value | 1.56551e-161 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
ZDHC9_MOUSE
|
||||||
| NC score | 0.924959 (rank : 5) | θ value | 9.16162e-53 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59268, Q5BL19 | Gene names | Zdhhc9 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9). | |||||
|
ZDHC9_HUMAN
|
||||||
| NC score | 0.924738 (rank : 6) | θ value | 9.16162e-53 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y397, Q59EK4, Q5JSW5, Q8WWS7, Q9BPY4, Q9NSP0, Q9NVL0, Q9NVR6 | Gene names | ZDHHC9, ZNF379 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9) (Zinc finger protein 379). | |||||
|
ZDH18_HUMAN
|
||||||
| NC score | 0.923114 (rank : 7) | θ value | 2.85459e-46 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NUE0, Q5JYH0, Q9H020 | Gene names | ZDHHC18 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC18 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 18) (DHHC-18). | |||||
|
ZDH14_MOUSE
|
||||||
| NC score | 0.913243 (rank : 8) | θ value | 3.98324e-48 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BQQ1, Q8BNR2, Q8CFN0 | Gene names | Zdhhc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
ZDH14_HUMAN
|
||||||
| NC score | 0.912224 (rank : 9) | θ value | 3.98324e-48 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IZN3, Q5JS07, Q5JS08, Q6PHS4, Q8IZN2, Q9H7F1 | Gene names | ZDHHC14 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
ZDH19_HUMAN
|
||||||
| NC score | 0.909078 (rank : 10) | θ value | 2.19584e-30 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVZ1 | Gene names | ZDHHC19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC19 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 19) (DHHC-19). | |||||
|
ZDH19_MOUSE
|
||||||
| NC score | 0.895848 (rank : 11) | θ value | 3.50572e-28 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q810M5 | Gene names | Zdhhc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC19 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 19) (DHHC-19). | |||||
|
ZDH12_MOUSE
|
||||||
| NC score | 0.850547 (rank : 12) | θ value | 1.52774e-15 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VC90, Q9CZ24, Q9D0T6 | Gene names | Zdhhc12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC12 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 12) (DHHC-12). | |||||
|
ZDH12_HUMAN
|
||||||
| NC score | 0.842884 (rank : 13) | θ value | 2.60593e-15 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96GR4, Q5T265, Q5T267, Q5T268, Q86VT5, Q96T09 | Gene names | ZDHHC12, ZNF400 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC12 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 12) (DHHC-12) (Zinc finger protein 400). | |||||
|
ZDHC4_MOUSE
|
||||||
| NC score | 0.826766 (rank : 14) | θ value | 8.10077e-17 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D6H5, Q8R5E0 | Gene names | Zdhhc4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC4 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 4) (DHHC-4). | |||||
|
ZDHC4_HUMAN
|
||||||
| NC score | 0.824590 (rank : 15) | θ value | 1.80466e-16 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NPG8, Q53EV7, Q6FIB5, Q9H0R9 | Gene names | ZDHHC4, ZNF374 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC4 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 4) (DHHC-4) (Zinc finger protein 374). | |||||
|
ZDH15_HUMAN
|
||||||
| NC score | 0.809290 (rank : 16) | θ value | 5.42112e-21 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96MV8, Q3SY30, Q6UWH3 | Gene names | ZDHHC15 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
ZDH15_MOUSE
|
||||||
| NC score | 0.808407 (rank : 17) | θ value | 5.42112e-21 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BGJ0, Q3TV63, Q8BMB7 | Gene names | Zdhhc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
ZDH16_MOUSE
|
||||||
| NC score | 0.805311 (rank : 18) | θ value | 4.02038e-16 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ESG8, Q3TI22, Q3UA59, Q91XC5 | Gene names | Zdhhc16, Aph2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC16 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 16) (DHHC-16) (Ablphilin-2). | |||||
|
ZDH20_HUMAN
|
||||||
| NC score | 0.805096 (rank : 19) | θ value | 4.43474e-23 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5W0Z9, Q6NVU8 | Gene names | ZDHHC20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC20 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 20) (DHHC-20). | |||||
|
ZDHC3_MOUSE
|
||||||
| NC score | 0.804842 (rank : 20) | θ value | 1.47631e-18 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8R173, Q6EMK3 | Gene names | Zdhhc3, Godz, Gramp1 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC3 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 3) (DHHC-3) (Golgi-specific DHHC zinc finger protein) (GABA-A receptor-associated membrane protein 1). | |||||
|
ZDHC3_HUMAN
|
||||||
| NC score | 0.799430 (rank : 21) | θ value | 1.24977e-17 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NYG2, Q53A17, Q96BL0 | Gene names | ZDHHC3, ZNF373 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC3 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 3) (DHHC-3) (Zinc finger protein 373) (DHHC1 protein). | |||||
|
ZDH20_MOUSE
|
||||||
| NC score | 0.798687 (rank : 22) | θ value | 3.51386e-20 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5Y5T1, Q3TDL1, Q8VCL6, Q9D3Q8 | Gene names | Zdhhc20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC20 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 20) (DHHC-20). | |||||
|
ZDHC2_MOUSE
|
||||||
| NC score | 0.795974 (rank : 23) | θ value | 2.06002e-20 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59267 | Gene names | Zdhhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC2 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 2) (DHHC-2). | |||||
|
ZDH16_HUMAN
|
||||||
| NC score | 0.795297 (rank : 24) | θ value | 1.68911e-14 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q969W1, Q5JTG7, Q5JTH0, Q8N4Z6, Q8WY84, Q9BSV3 | Gene names | ZDHHC16 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC16 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 16) (DHHC-16). | |||||
|
ZDHC2_HUMAN
|
||||||
| NC score | 0.795196 (rank : 25) | θ value | 2.06002e-20 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UIJ5 | Gene names | ZDHHC2, REAM, REC, ZNF372 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC2 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 2) (DHHC-2) (Zinc finger protein 372) (Reduced expression associated with metastasis protein) (Ream) (Reduced expression in cancer protein) (Rec). | |||||
|
ZDHC6_MOUSE
|
||||||
| NC score | 0.793103 (rank : 26) | θ value | 5.25075e-16 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CPV7, Q3U3I8, Q3UT70, Q544H1 | Gene names | Zdhhc6 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC6 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 6) (DHHC-6) (H4 homolog). | |||||
|
ZDH21_HUMAN
|
||||||
| NC score | 0.792896 (rank : 27) | θ value | 3.52202e-12 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IVQ6, Q5VWG1 | Gene names | ZDHHC21 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC21 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 21) (DHHC-21). | |||||
|
ZDHC6_HUMAN
|
||||||
| NC score | 0.791121 (rank : 28) | θ value | 5.25075e-16 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H6R6, Q53G45, Q96IV7, Q9H605 | Gene names | ZDHHC6, ZNF376 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC6 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 6) (DHHC-6) (Zinc finger protein 376) (Transmembrane protein H4). | |||||
|
ZDH21_MOUSE
|
||||||
| NC score | 0.788821 (rank : 29) | θ value | 1.74796e-11 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D270, Q6EMK1, Q80XQ3 | Gene names | Zdhhc21, Gramp3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC21 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 21) (DHHC-21) (GABA-A receptor-associated membrane protein 3). | |||||
|
ZDHC7_MOUSE
|
||||||
| NC score | 0.787574 (rank : 30) | θ value | 4.02038e-16 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91WU6, Q3TC41, Q6EMK2, Q8K1H6 | Gene names | Zdhhc7, Gramp2 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC7 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 7) (DHHC-7) (GABA-A receptor-associated membrane protein 2). | |||||
|
ZDHC7_HUMAN
|
||||||
| NC score | 0.787458 (rank : 31) | θ value | 1.058e-16 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NXF8, Q8WV42, Q9NVD8 | Gene names | ZDHHC7, ZNF370 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC7 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 7) (DHHC-7) (Zinc finger protein 370). | |||||
|
ZDH11_HUMAN
|
||||||
| NC score | 0.785159 (rank : 32) | θ value | 4.16044e-13 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H8X9 | Gene names | ZDHHC11, ZNF399 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC11 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 11) (DHHC-11) (Zinc finger protein 399). | |||||
|
ZDH24_HUMAN
|
||||||
| NC score | 0.776996 (rank : 33) | θ value | 2.52405e-10 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6UX98, Q6PEW7, Q9BSJ0 | Gene names | ZDHHC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC24 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 24) (DHHC-24). | |||||
|
ZDH24_MOUSE
|
||||||
| NC score | 0.769757 (rank : 34) | θ value | 2.52405e-10 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6IR37, Q3TAY4, Q3TDF1, Q3TF09, Q9CS66 | Gene names | Zdhhc24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC24 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 24) (DHHC-24). | |||||
|
ZDH23_MOUSE
|
||||||
| NC score | 0.768476 (rank : 35) | θ value | 2.69671e-12 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5Y5T3, Q3UH65, Q3UVA2 | Gene names | Zdhhc23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC23 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 23) (DHHC-23) (DHHC-containing protein 11). | |||||
|
ZDH23_HUMAN
|
||||||
| NC score | 0.748681 (rank : 36) | θ value | 2.98157e-11 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IYP9 | Gene names | ZDHHC23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC23 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 23) (DHHC-23). | |||||
|
ZDHC1_MOUSE
|
||||||
| NC score | 0.742022 (rank : 37) | θ value | 3.89403e-11 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8R0N9, Q8BJ24 | Gene names | Zdhhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC1 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 1) (DHHC-1). | |||||
|
ZDHC1_HUMAN
|
||||||
| NC score | 0.737231 (rank : 38) | θ value | 2.28291e-11 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WTX9, O15461 | Gene names | ZDHHC1, C16orf1, ZNF377 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC1 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 1) (DHHC-1) (Zinc finger protein 377) (DHHC- domain-containing cysteine-rich protein 1). | |||||
|
ZDH22_HUMAN
|
||||||
| NC score | 0.712032 (rank : 39) | θ value | 0.000786445 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N966 | Gene names | ZDHHC22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative palmitoyltransferase ZDHHC22 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 22) (DHHC-22). | |||||
|
ZDH17_MOUSE
|
||||||
| NC score | 0.453012 (rank : 40) | θ value | 8.65492e-19 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
ZDH17_HUMAN
|
||||||
| NC score | 0.447774 (rank : 41) | θ value | 6.62687e-19 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
ZDH13_MOUSE
|
||||||
| NC score | 0.436033 (rank : 42) | θ value | 3.52202e-12 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9CWU2, Q3UK32, Q3UKV1 | Gene names | Zdhhc13, Hip14l | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein). | |||||
|
ZDH13_HUMAN
|
||||||
| NC score | 0.430432 (rank : 43) | θ value | 2.28291e-11 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8IUH4, Q7Z2D3, Q86VK2, Q9NV30, Q9NV99 | Gene names | ZDHHC13, HIP14L, HIP3RP | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein) (Huntingtin- interacting protein HIP3RP) (Putative NF-kappa-B-activating protein 209) (Putative MAPK-activating protein PM03). | |||||
|
ZCHC4_HUMAN
|
||||||
| NC score | 0.112223 (rank : 44) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H5U6, Q5IW78, Q96AN7 | Gene names | ZCCHC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
ZCHC4_MOUSE
|
||||||
| NC score | 0.082485 (rank : 45) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKW4, Q3UTX6, Q9D2I1 | Gene names | Zcchc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.060124 (rank : 46) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PRPE_HUMAN
|
||||||
| NC score | 0.056666 (rank : 47) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.051376 (rank : 48) | θ value | 0.0431538 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.050986 (rank : 49) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.050842 (rank : 50) | θ value | 0.0431538 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.045749 (rank : 51) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.045485 (rank : 52) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
AKTS1_MOUSE
|
||||||
| NC score | 0.042343 (rank : 53) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D1F4 | Gene names | Akt1s1, Pras | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (Proline-rich AKT substrate). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.040362 (rank : 54) | θ value | 0.0252991 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
AKTS1_HUMAN
|
||||||
| NC score | 0.039312 (rank : 55) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96B36, Q96BI4, Q96IK7, Q96NG2, Q9BWR5 | Gene names | AKT1S1, PRAS40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (40 kDa proline-rich AKT substrate). | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.034129 (rank : 56) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.030495 (rank : 57) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.030224 (rank : 58) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
LPIN1_MOUSE
|
||||||
| NC score | 0.029372 (rank : 59) | θ value | 0.21417 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZP3, Q9CQI2, Q9JLG6 | Gene names | Lpin1, Fld | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-1 (Fatty liver dystrophy protein). | |||||
|
IL16_HUMAN
|
||||||
| NC score | 0.028910 (rank : 60) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.028253 (rank : 61) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.028008 (rank : 62) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.027976 (rank : 63) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.027943 (rank : 64) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.027931 (rank : 65) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.027802 (rank : 66) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.027595 (rank : 67) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
IP3KB_HUMAN
|
||||||
| NC score | 0.027491 (rank : 68) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.027306 (rank : 69) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.027148 (rank : 70) | θ value | 0.163984 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.026721 (rank : 71) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
GAK3_HUMAN
|
||||||
| NC score | 0.026617 (rank : 72) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.026425 (rank : 73) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.026075 (rank : 74) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.024454 (rank : 75) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.024159 (rank : 76) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.021876 (rank : 77) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.021315 (rank : 78) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
LPIN3_MOUSE
|
||||||
| NC score | 0.020882 (rank : 79) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-3. | |||||
|
PHLB1_HUMAN
|
||||||
| NC score | 0.018452 (rank : 80) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
OCLN_MOUSE
|
||||||
| NC score | 0.016560 (rank : 81) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61146, Q544A7 | Gene names | Ocln, Ocl | |||
|
Domain Architecture |

|
|||||
| Description | Occludin. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.016481 (rank : 82) | θ value | 0.0736092 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.016262 (rank : 83) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.015957 (rank : 84) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.015737 (rank : 85) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
K0562_MOUSE
|
||||||
| NC score | 0.015621 (rank : 86) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80V31, Q6ZQ95 | Gene names | Kiaa0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
THUM1_MOUSE
|
||||||
| NC score | 0.015421 (rank : 87) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J36, Q3U6Y3 | Gene names | Thumpd1 | |||
|
Domain Architecture |

|
|||||
| Description | THUMP domain-containing protein 1. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.014624 (rank : 88) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.013647 (rank : 89) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.013267 (rank : 90) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.011523 (rank : 91) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.010456 (rank : 92) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.008968 (rank : 93) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.008635 (rank : 94) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.008550 (rank : 95) | θ value | 0.21417 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.008097 (rank : 96) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
TTLL4_MOUSE
|
||||||
| NC score | 0.007400 (rank : 97) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
SEM4A_HUMAN
|
||||||
| NC score | 0.007094 (rank : 98) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
AMOL2_MOUSE
|
||||||
| NC score | 0.006750 (rank : 99) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
UBQL3_MOUSE
|
||||||
| NC score | 0.006724 (rank : 100) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5U9 | Gene names | Ubqln3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.006374 (rank : 101) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
SLIK3_HUMAN
|
||||||
| NC score | 0.004821 (rank : 102) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94933 | Gene names | SLITRK3, KIAA0848 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLIT and NTRK-like protein 3 precursor. | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.004472 (rank : 103) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.003921 (rank : 104) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NOS1_MOUSE
|
||||||
| NC score | 0.003892 (rank : 105) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0J4, Q64208 | Gene names | Nos1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
SEM6C_HUMAN
|
||||||
| NC score | 0.003151 (rank : 106) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.002273 (rank : 107) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
HME1_MOUSE
|
||||||
| NC score | 0.001326 (rank : 108) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09065 | Gene names | En1, En-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein engrailed-1 (Mo-En-1). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | -0.000136 (rank : 109) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | -0.001441 (rank : 110) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
O51D1_HUMAN
|
||||||
| NC score | -0.002150 (rank : 111) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGF3 | Gene names | OR51D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 51D1 (Olfactory receptor OR11-14). | |||||
|
CLK3_HUMAN
|
||||||
| NC score | -0.002613 (rank : 112) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49761, Q9BRS3, Q9BUJ7 | Gene names | CLK3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase CLK3 (EC 2.7.12.1) (CDC-like kinase 3). | |||||
|
MLTK_HUMAN
|
||||||
| NC score | -0.002862 (rank : 113) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
DUET_HUMAN
|
||||||
| NC score | -0.003527 (rank : 114) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||
|
GLI3_MOUSE
|
||||||
| NC score | -0.004607 (rank : 115) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||