Please be patient as the page loads
|
GCYA2_HUMAN
|
||||||
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GCYA2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
GCYA3_HUMAN
|
||||||
| θ value | 8.00302e-174 (rank : 2) | NC score | 0.990726 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02108, O43843, Q8TAH3 | Gene names | GUCY1A3, GUC1A3, GUCSA3, GUCY1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYA3_MOUSE
|
||||||
| θ value | 1.2777e-171 (rank : 3) | NC score | 0.990095 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ERL9, Q9DBQ3 | Gene names | Gucy1a3, Gucy1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYB1_MOUSE
|
||||||
| θ value | 7.66637e-92 (rank : 4) | NC score | 0.968924 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54865 | Gene names | Gucy1b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit). | |||||
|
GCYB1_HUMAN
|
||||||
| θ value | 1.00126e-91 (rank : 5) | NC score | 0.968650 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02153, Q86WY5 | Gene names | GUCY1B3, GUC1B3, GUCSB3, GUCY1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit) (GCS-beta-3). | |||||
|
GCYB2_HUMAN
|
||||||
| θ value | 3.23591e-74 (rank : 6) | NC score | 0.967677 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75343, Q9NZ64 | Gene names | GUCY1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-2 (EC 4.6.1.2) (GCS-beta-2). | |||||
|
ANPRA_MOUSE
|
||||||
| θ value | 7.51208e-47 (rank : 7) | NC score | 0.539095 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRB_HUMAN
|
||||||
| θ value | 1.85029e-45 (rank : 8) | NC score | 0.532576 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRB_MOUSE
|
||||||
| θ value | 4.122e-45 (rank : 9) | NC score | 0.549325 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRA_HUMAN
|
||||||
| θ value | 7.03111e-45 (rank : 10) | NC score | 0.538068 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
GUC2E_MOUSE
|
||||||
| θ value | 3.85808e-43 (rank : 11) | NC score | 0.594484 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
GUC2G_MOUSE
|
||||||
| θ value | 4.71619e-41 (rank : 12) | NC score | 0.551795 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
GUC2C_HUMAN
|
||||||
| θ value | 1.79215e-40 (rank : 13) | NC score | 0.519957 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
GUC2F_HUMAN
|
||||||
| θ value | 4.41421e-39 (rank : 14) | NC score | 0.501177 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
GUC2D_HUMAN
|
||||||
| θ value | 3.73686e-38 (rank : 15) | NC score | 0.591420 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
ADCY1_HUMAN
|
||||||
| θ value | 3.28125e-26 (rank : 16) | NC score | 0.801653 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08828, Q75MI1 | Gene names | ADCY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY1_MOUSE
|
||||||
| θ value | 3.28125e-26 (rank : 17) | NC score | 0.799014 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88444, Q5SS89 | Gene names | Adcy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY2_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 18) | NC score | 0.785663 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08462, Q9UPU2 | Gene names | ADCY2, KIAA1060 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY2_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 19) | NC score | 0.782893 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80TL1, Q8CFU6 | Gene names | Adcy2, Kiaa1060 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY6_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 20) | NC score | 0.790606 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43306, Q9NR75 | Gene names | ADCY6, KIAA0422 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY6_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 21) | NC score | 0.788313 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01341 | Gene names | Adcy6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY7_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 22) | NC score | 0.789675 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51829 | Gene names | Adcy7 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY4_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 23) | NC score | 0.788260 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFM4, Q96ML7 | Gene names | ADCY4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY4_MOUSE
|
||||||
| θ value | 6.84181e-24 (rank : 24) | NC score | 0.790227 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91WF3 | Gene names | Adcy4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY3_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 25) | NC score | 0.794950 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY3_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 26) | NC score | 0.796975 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VHH7 | Gene names | Adcy3 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY7_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 27) | NC score | 0.788122 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51828 | Gene names | ADCY7, KIAA0037 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY8_HUMAN
|
||||||
| θ value | 4.9032e-22 (rank : 28) | NC score | 0.784124 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40145 | Gene names | ADCY8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY8_MOUSE
|
||||||
| θ value | 6.40375e-22 (rank : 29) | NC score | 0.783583 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97490 | Gene names | Adcy8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY9_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 30) | NC score | 0.803760 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60503, O60273, Q4ZHT9, Q4ZIR5, Q9BWT4, Q9UGP2 | Gene names | ADCY9, KIAA0520 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9). | |||||
|
ADCY9_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 31) | NC score | 0.803301 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51830, Q61279 | Gene names | Adcy9 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9) (Adenylyl cyclase type 10) (ACTP10). | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.007732 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
KIF17_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.007562 (rank : 63) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2E2, O95077, Q53YS6, Q5VWA9, Q6GSA8, Q8N411 | Gene names | KIF17, KIAA1405, KIF3X | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF17 (KIF3-related motor protein). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.016273 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
PSA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.022079 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25788, Q86U83, Q8N1D8, Q9BS70 | Gene names | PSMA3, PSC8 | |||
|
Domain Architecture |
|
|||||
| Description | Proteasome subunit alpha type 3 (EC 3.4.25.1) (Proteasome component C8) (Macropain subunit C8) (Multicatalytic endopeptidase complex subunit C8). | |||||
|
PSA3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.022044 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70435 | Gene names | Psma3 | |||
|
Domain Architecture |
|
|||||
| Description | Proteasome subunit alpha type 3 (EC 3.4.25.1) (Proteasome component C8) (Macropain subunit C8) (Multicatalytic endopeptidase complex subunit C8) (Proteasome subunit K). | |||||
|
CENPT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.015204 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.003105 (rank : 69) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
BAG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.014519 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99933, O75315, Q14414, Q9Y2V4 | Gene names | BAG1 | |||
|
Domain Architecture |
|
|||||
| Description | BAG family molecular chaperone regulator 1 (BCL-2-binding athanogene- 1) (BAG-1) (Glucocorticoid receptor-associated protein RAP46). | |||||
|
LIFR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.007256 (rank : 64) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42703 | Gene names | Lifr | |||
|
Domain Architecture |

|
|||||
| Description | Leukemia inhibitory factor receptor precursor (LIF receptor) (LIF-R) (D-factor/LIF receptor) (CD118 antigen). | |||||
|
LZTS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.006537 (rank : 66) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.000584 (rank : 74) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
PRIC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.006880 (rank : 65) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80Y24 | Gene names | Prickle2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | -0.000225 (rank : 75) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
NMD3B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.003953 (rank : 68) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
PRIC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.006388 (rank : 67) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z3G6 | Gene names | PRICKLE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
FOXE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.002080 (rank : 71) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.002808 (rank : 70) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.000922 (rank : 73) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.001513 (rank : 72) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
ANPRC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.267002 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
ANPRC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.260992 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
GABR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.074837 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.077133 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
ILK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.059226 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13418 | Gene names | ILK, ILK1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase 1 (EC 2.7.11.1) (ILK-1) (59 kDa serine/threonine-protein kinase) (p59ILK). | |||||
|
ILK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.059500 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O55222 | Gene names | Ilk | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase (EC 2.7.11.1). | |||||
|
IRAK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.071734 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y616 | Gene names | IRAK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
IRAK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.069589 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4B2, Q8C7U8, Q8CE40, Q8K1S8 | Gene names | Irak3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
KSR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.069780 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
KSR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.068789 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
KSR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.062541 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6VAB6, Q8N775 | Gene names | KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-2 (hKSR2). | |||||
|
MLKL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.069241 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NB16, Q8N6V0 | Gene names | MLKL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
MLKL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.068268 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D2Y4, Q8CIJ5 | Gene names | Mlkl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
MLTK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.051255 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
MLTK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.051566 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESL4, Q3V1X8, Q8BR73, Q9ESL3 | Gene names | Mltk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif kinase ZAK) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK). | |||||
|
RIPK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.059661 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13546, Q13180 | Gene names | RIPK1, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
RIPK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.061769 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60855, Q3U0J3, Q8CD90 | Gene names | Ripk1, Rinp, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
RIPK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.071447 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43353, Q6UWF0 | Gene names | RIPK2, CARDIAK, RICK, RIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (RIP-like-interacting CLARP kinase) (Receptor-interacting protein 2) (RIP-2) (CARD-containing interleukin-1 beta-converting enzyme- associated kinase) (CARD-containing IL-1 beta ICE-kinase). | |||||
|
RIPK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.069868 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58801 | Gene names | Ripk2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
RIPK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.051653 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y572 | Gene names | RIPK3, RIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3). | |||||
|
RIPK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.052943 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |
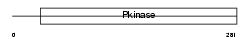
|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
TESK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.050070 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96S53, Q8N520, Q9Y3Q6 | Gene names | TESK2 | |||
|
Domain Architecture |
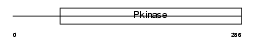
|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TESK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050140 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |
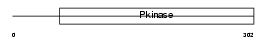
|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TOPK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.055793 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KB5, Q9NPD9, Q9NYL7, Q9NZK6 | Gene names | PBK, TOPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase) (Spermatogenesis-related protein kinase) (SPK) (MAPKK-like protein kinase) (Nori-3). | |||||
|
TOPK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.053200 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJ78, Q922V2, Q9D184 | Gene names | Pbk, Topk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase). | |||||
|
GCYA2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
GCYA3_HUMAN
|
||||||
| NC score | 0.990726 (rank : 2) | θ value | 8.00302e-174 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02108, O43843, Q8TAH3 | Gene names | GUCY1A3, GUC1A3, GUCSA3, GUCY1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYA3_MOUSE
|
||||||
| NC score | 0.990095 (rank : 3) | θ value | 1.2777e-171 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ERL9, Q9DBQ3 | Gene names | Gucy1a3, Gucy1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYB1_MOUSE
|
||||||
| NC score | 0.968924 (rank : 4) | θ value | 7.66637e-92 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54865 | Gene names | Gucy1b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit). | |||||
|
GCYB1_HUMAN
|
||||||
| NC score | 0.968650 (rank : 5) | θ value | 1.00126e-91 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02153, Q86WY5 | Gene names | GUCY1B3, GUC1B3, GUCSB3, GUCY1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit) (GCS-beta-3). | |||||
|
GCYB2_HUMAN
|
||||||
| NC score | 0.967677 (rank : 6) | θ value | 3.23591e-74 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75343, Q9NZ64 | Gene names | GUCY1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-2 (EC 4.6.1.2) (GCS-beta-2). | |||||
|
ADCY9_HUMAN
|
||||||
| NC score | 0.803760 (rank : 7) | θ value | 2.06002e-20 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60503, O60273, Q4ZHT9, Q4ZIR5, Q9BWT4, Q9UGP2 | Gene names | ADCY9, KIAA0520 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9). | |||||
|
ADCY9_MOUSE
|
||||||
| NC score | 0.803301 (rank : 8) | θ value | 2.06002e-20 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51830, Q61279 | Gene names | Adcy9 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9) (Adenylyl cyclase type 10) (ACTP10). | |||||
|
ADCY1_HUMAN
|
||||||
| NC score | 0.801653 (rank : 9) | θ value | 3.28125e-26 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08828, Q75MI1 | Gene names | ADCY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY1_MOUSE
|
||||||
| NC score | 0.799014 (rank : 10) | θ value | 3.28125e-26 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88444, Q5SS89 | Gene names | Adcy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY3_MOUSE
|
||||||
| NC score | 0.796975 (rank : 11) | θ value | 1.16704e-23 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VHH7 | Gene names | Adcy3 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY3_HUMAN
|
||||||
| NC score | 0.794950 (rank : 12) | θ value | 1.16704e-23 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY6_HUMAN
|
||||||
| NC score | 0.790606 (rank : 13) | θ value | 2.12685e-25 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43306, Q9NR75 | Gene names | ADCY6, KIAA0422 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY4_MOUSE
|
||||||
| NC score | 0.790227 (rank : 14) | θ value | 6.84181e-24 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91WF3 | Gene names | Adcy4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY7_MOUSE
|
||||||
| NC score | 0.789675 (rank : 15) | θ value | 4.01107e-24 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51829 | Gene names | Adcy7 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY6_MOUSE
|
||||||
| NC score | 0.788313 (rank : 16) | θ value | 1.05554e-24 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01341 | Gene names | Adcy6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY4_HUMAN
|
||||||
| NC score | 0.788260 (rank : 17) | θ value | 6.84181e-24 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFM4, Q96ML7 | Gene names | ADCY4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY7_HUMAN
|
||||||
| NC score | 0.788122 (rank : 18) | θ value | 1.5242e-23 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51828 | Gene names | ADCY7, KIAA0037 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY2_HUMAN
|
||||||
| NC score | 0.785663 (rank : 19) | θ value | 7.30988e-26 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08462, Q9UPU2 | Gene names | ADCY2, KIAA1060 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY8_HUMAN
|
||||||
| NC score | 0.784124 (rank : 20) | θ value | 4.9032e-22 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40145 | Gene names | ADCY8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY8_MOUSE
|
||||||
| NC score | 0.783583 (rank : 21) | θ value | 6.40375e-22 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97490 | Gene names | Adcy8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY2_MOUSE
|
||||||
| NC score | 0.782893 (rank : 22) | θ value | 7.30988e-26 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80TL1, Q8CFU6 | Gene names | Adcy2, Kiaa1060 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
GUC2E_MOUSE
|
||||||
| NC score | 0.594484 (rank : 23) | θ value | 3.85808e-43 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
GUC2D_HUMAN
|
||||||
| NC score | 0.591420 (rank : 24) | θ value | 3.73686e-38 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
GUC2G_MOUSE
|
||||||
| NC score | 0.551795 (rank : 25) | θ value | 4.71619e-41 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
ANPRB_MOUSE
|
||||||
| NC score | 0.549325 (rank : 26) | θ value | 4.122e-45 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRA_MOUSE
|
||||||
| NC score | 0.539095 (rank : 27) | θ value | 7.51208e-47 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRA_HUMAN
|
||||||
| NC score | 0.538068 (rank : 28) | θ value | 7.03111e-45 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRB_HUMAN
|
||||||
| NC score | 0.532576 (rank : 29) | θ value | 1.85029e-45 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
GUC2C_HUMAN
|
||||||
| NC score | 0.519957 (rank : 30) | θ value | 1.79215e-40 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
GUC2F_HUMAN
|
||||||
| NC score | 0.501177 (rank : 31) | θ value | 4.41421e-39 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
ANPRC_HUMAN
|
||||||
| NC score | 0.267002 (rank : 32) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
ANPRC_MOUSE
|
||||||
| NC score | 0.260992 (rank : 33) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.077133 (rank : 34) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.074837 (rank : 35) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
IRAK3_HUMAN
|
||||||
| NC score | 0.071734 (rank : 36) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y616 | Gene names | IRAK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
RIPK2_HUMAN
|
||||||
| NC score | 0.071447 (rank : 37) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43353, Q6UWF0 | Gene names | RIPK2, CARDIAK, RICK, RIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (RIP-like-interacting CLARP kinase) (Receptor-interacting protein 2) (RIP-2) (CARD-containing interleukin-1 beta-converting enzyme- associated kinase) (CARD-containing IL-1 beta ICE-kinase). | |||||
|
RIPK2_MOUSE
|
||||||
| NC score | 0.069868 (rank : 38) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58801 | Gene names | Ripk2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
KSR1_HUMAN
|
||||||
| NC score | 0.069780 (rank : 39) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
IRAK3_MOUSE
|
||||||
| NC score | 0.069589 (rank : 40) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4B2, Q8C7U8, Q8CE40, Q8K1S8 | Gene names | Irak3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
MLKL_HUMAN
|
||||||
| NC score | 0.069241 (rank : 41) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NB16, Q8N6V0 | Gene names | MLKL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
KSR1_MOUSE
|
||||||
| NC score | 0.068789 (rank : 42) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
MLKL_MOUSE
|
||||||
| NC score | 0.068268 (rank : 43) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D2Y4, Q8CIJ5 | Gene names | Mlkl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
KSR2_HUMAN
|
||||||
| NC score | 0.062541 (rank : 44) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6VAB6, Q8N775 | Gene names | KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-2 (hKSR2). | |||||
|
RIPK1_MOUSE
|
||||||
| NC score | 0.061769 (rank : 45) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60855, Q3U0J3, Q8CD90 | Gene names | Ripk1, Rinp, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
RIPK1_HUMAN
|
||||||
| NC score | 0.059661 (rank : 46) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13546, Q13180 | Gene names | RIPK1, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
ILK_MOUSE
|
||||||
| NC score | 0.059500 (rank : 47) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O55222 | Gene names | Ilk | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase (EC 2.7.11.1). | |||||
|
ILK1_HUMAN
|
||||||
| NC score | 0.059226 (rank : 48) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13418 | Gene names | ILK, ILK1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase 1 (EC 2.7.11.1) (ILK-1) (59 kDa serine/threonine-protein kinase) (p59ILK). | |||||
|
TOPK_HUMAN
|
||||||
| NC score | 0.055793 (rank : 49) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KB5, Q9NPD9, Q9NYL7, Q9NZK6 | Gene names | PBK, TOPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase) (Spermatogenesis-related protein kinase) (SPK) (MAPKK-like protein kinase) (Nori-3). | |||||
|
TOPK_MOUSE
|
||||||
| NC score | 0.053200 (rank : 50) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJ78, Q922V2, Q9D184 | Gene names | Pbk, Topk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase). | |||||
|
RIPK3_MOUSE
|
||||||
| NC score | 0.052943 (rank : 51) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
RIPK3_HUMAN
|
||||||
| NC score | 0.051653 (rank : 52) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y572 | Gene names | RIPK3, RIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3). | |||||
|
MLTK_MOUSE
|
||||||
| NC score | 0.051566 (rank : 53) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESL4, Q3V1X8, Q8BR73, Q9ESL3 | Gene names | Mltk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif kinase ZAK) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK). | |||||
|
MLTK_HUMAN
|
||||||
| NC score | 0.051255 (rank : 54) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
TESK2_MOUSE
|
||||||
| NC score | 0.050140 (rank : 55) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TESK2_HUMAN
|
||||||
| NC score | 0.050070 (rank : 56) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96S53, Q8N520, Q9Y3Q6 | Gene names | TESK2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
PSA3_HUMAN
|
||||||
| NC score | 0.022079 (rank : 57) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25788, Q86U83, Q8N1D8, Q9BS70 | Gene names | PSMA3, PSC8 | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit alpha type 3 (EC 3.4.25.1) (Proteasome component C8) (Macropain subunit C8) (Multicatalytic endopeptidase complex subunit C8). | |||||
|
PSA3_MOUSE
|
||||||
| NC score | 0.022044 (rank : 58) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70435 | Gene names | Psma3 | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit alpha type 3 (EC 3.4.25.1) (Proteasome component C8) (Macropain subunit C8) (Multicatalytic endopeptidase complex subunit C8) (Proteasome subunit K). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.016273 (rank : 59) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
CENPT_MOUSE
|
||||||
| NC score | 0.015204 (rank : 60) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
BAG1_HUMAN
|
||||||
| NC score | 0.014519 (rank : 61) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99933, O75315, Q14414, Q9Y2V4 | Gene names | BAG1 | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 1 (BCL-2-binding athanogene- 1) (BAG-1) (Glucocorticoid receptor-associated protein RAP46). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.007732 (rank : 62) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
KIF17_HUMAN
|
||||||
| NC score | 0.007562 (rank : 63) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2E2, O95077, Q53YS6, Q5VWA9, Q6GSA8, Q8N411 | Gene names | KIF17, KIAA1405, KIF3X | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (KIF3-related motor protein). | |||||
|
LIFR_MOUSE
|
||||||
| NC score | 0.007256 (rank : 64) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42703 | Gene names | Lifr | |||
|
Domain Architecture |

|
|||||
| Description | Leukemia inhibitory factor receptor precursor (LIF receptor) (LIF-R) (D-factor/LIF receptor) (CD118 antigen). | |||||
|
PRIC2_MOUSE
|
||||||
| NC score | 0.006880 (rank : 65) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80Y24 | Gene names | Prickle2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
LZTS2_MOUSE
|
||||||
| NC score | 0.006537 (rank : 66) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
PRIC2_HUMAN
|
||||||
| NC score | 0.006388 (rank : 67) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z3G6 | Gene names | PRICKLE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
NMD3B_HUMAN
|
||||||
| NC score | 0.003953 (rank : 68) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.003105 (rank : 69) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.002808 (rank : 70) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
FOXE2_HUMAN
|
||||||
| NC score | 0.002080 (rank : 71) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.001513 (rank : 72) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.000922 (rank : 73) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.000584 (rank : 74) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | -0.000225 (rank : 75) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||