Please be patient as the page loads
|
SYV_MOUSE
|
||||||
| SwissProt Accessions | Q9Z1Q9, Q9QUN2 | Gene names | Vars, Bat6, G7a, Vars2 | |||
|
Domain Architecture |
|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SYV_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994258 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26640, Q5JQ90, Q96E77, Q9UQM2 | Gene names | VARS, G7A, VARS2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
SYV_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z1Q9, Q9QUN2 | Gene names | Vars, Bat6, G7a, Vars2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
SYIM_MOUSE
|
||||||
| θ value | 1.72781e-51 (rank : 3) | NC score | 0.762480 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BIJ6, Q3TKT8, Q8BIJ0, Q8BIP3 | Gene names | Iars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
SYIM_HUMAN
|
||||||
| θ value | 6.7944e-48 (rank : 4) | NC score | 0.756982 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NSE4, Q6PI85, Q7L439, Q86WU9, Q96D91 | Gene names | IARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
SYIC_HUMAN
|
||||||
| θ value | 2.05049e-36 (rank : 5) | NC score | 0.694356 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P41252, Q5TCD0, Q9H588 | Gene names | IARS | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
SYIC_MOUSE
|
||||||
| θ value | 2.50655e-34 (rank : 6) | NC score | 0.692172 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
EF1G_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 7) | NC score | 0.552033 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
EF1G_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 8) | NC score | 0.550959 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
SYLM_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 9) | NC score | 0.485174 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15031 | Gene names | LARS2, KIAA0028 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLM_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 10) | NC score | 0.480554 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VDC0 | Gene names | Lars2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYMM_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 11) | NC score | 0.241417 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
SYMM_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 12) | NC score | 0.219756 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96GW9, Q76E79, Q8IW62, Q8N7N4 | Gene names | MARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
SYLC_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 13) | NC score | 0.229778 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMJ2, Q8BKW9 | Gene names | Lars | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLC_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 14) | NC score | 0.236033 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2J5, Q9NSE1 | Gene names | LARS, KIAA1352 | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYEP_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.079501 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SYM_MOUSE
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.106060 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
SYM_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.151583 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.008348 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
GSTT2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.066750 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30712, O60665, Q9HD76 | Gene names | GSTT2 | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
IBP5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.018224 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.023716 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
ACM3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.002276 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERZ3, Q64055 | Gene names | Chrm3, Chrm-3 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M3 (Mm3 mAChR). | |||||
|
MCA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.135944 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D1M4 | Gene names | Eef1e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.007533 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SYEP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.049778 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
ZN232_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | -0.000673 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNY5 | Gene names | ZNF232, ZSCAN11 | |||
|
Domain Architecture |
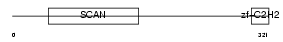
|
|||||
| Description | Zinc finger protein 232 (Zinc finger and SCAN domain-containing protein 11). | |||||
|
CNTP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.010835 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.015905 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.015914 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.014540 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.008407 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
ZAR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.019912 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
KS6B1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.002179 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23443 | Gene names | RPS6KB1 | |||
|
Domain Architecture |
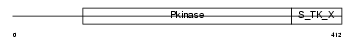
|
|||||
| Description | Ribosomal protein S6 kinase beta-1 (EC 2.7.11.1) (Ribosomal protein S6 kinase I) (S6K) (S6K1) (70 kDa ribosomal protein S6 kinase 1) (p70 S6 kinase alpha) (p70(S6K)-alpha) (p70-S6K) (P70S6K) (p70-alpha). | |||||
|
KS6B1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.002180 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BSK8, Q8CHX0 | Gene names | Rps6kb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase beta-1 (EC 2.7.11.1) (Ribosomal protein S6 kinase I) (S6K) (S6K1) (70 kDa ribosomal protein S6 kinase 1) (p70 S6 kinase alpha) (p70(S6K)-alpha) (p70-S6K) (P70S6K). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.012849 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
UBP14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.011711 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.002622 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
KS6B2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.001636 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBS0, O94809, Q9UEC1 | Gene names | RPS6KB2, STK14B | |||
|
Domain Architecture |
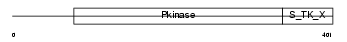
|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (p70 S6 kinase beta) (S6K-beta) (p70-beta) (S6 kinase-related kinase) (SRK) (Serine/threonine-protein kinase 14 beta). | |||||
|
NAF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.006677 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
PGTA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.010607 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHK4, Q9JLX2 | Gene names | Rabggta | |||
|
Domain Architecture |
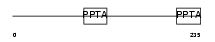
|
|||||
| Description | Geranylgeranyl transferase type-2 alpha subunit (EC 2.5.1.60) (Geranylgeranyl transferase type II alpha subunit) (Rab geranylgeranyltransferase alpha subunit) (Rab geranyl- geranyltransferase alpha subunit) (Rab GG transferase alpha) (Rab GGTase alpha). | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.009355 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
CN092_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.007767 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CNTP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.009068 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
DPOD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.011501 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
DYH9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.006159 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.001171 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TRPS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.011417 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
MCA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.083380 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43324, Q5THS2 | Gene names | EEF1E1 | |||
|
Domain Architecture |
|
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
SYV_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z1Q9, Q9QUN2 | Gene names | Vars, Bat6, G7a, Vars2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
SYV_HUMAN
|
||||||
| NC score | 0.994258 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26640, Q5JQ90, Q96E77, Q9UQM2 | Gene names | VARS, G7A, VARS2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
SYIM_MOUSE
|
||||||
| NC score | 0.762480 (rank : 3) | θ value | 1.72781e-51 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BIJ6, Q3TKT8, Q8BIJ0, Q8BIP3 | Gene names | Iars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
SYIM_HUMAN
|
||||||
| NC score | 0.756982 (rank : 4) | θ value | 6.7944e-48 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NSE4, Q6PI85, Q7L439, Q86WU9, Q96D91 | Gene names | IARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
SYIC_HUMAN
|
||||||
| NC score | 0.694356 (rank : 5) | θ value | 2.05049e-36 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P41252, Q5TCD0, Q9H588 | Gene names | IARS | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
SYIC_MOUSE
|
||||||
| NC score | 0.692172 (rank : 6) | θ value | 2.50655e-34 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
EF1G_HUMAN
|
||||||
| NC score | 0.552033 (rank : 7) | θ value | 5.59698e-26 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
EF1G_MOUSE
|
||||||
| NC score | 0.550959 (rank : 8) | θ value | 1.24688e-25 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
SYLM_HUMAN
|
||||||
| NC score | 0.485174 (rank : 9) | θ value | 1.86753e-13 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15031 | Gene names | LARS2, KIAA0028 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLM_MOUSE
|
||||||
| NC score | 0.480554 (rank : 10) | θ value | 3.52202e-12 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VDC0 | Gene names | Lars2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYMM_MOUSE
|
||||||
| NC score | 0.241417 (rank : 11) | θ value | 9.29e-05 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
SYLC_HUMAN
|
||||||
| NC score | 0.236033 (rank : 12) | θ value | 0.0193708 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2J5, Q9NSE1 | Gene names | LARS, KIAA1352 | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLC_MOUSE
|
||||||
| NC score | 0.229778 (rank : 13) | θ value | 0.00390308 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMJ2, Q8BKW9 | Gene names | Lars | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYMM_HUMAN
|
||||||
| NC score | 0.219756 (rank : 14) | θ value | 0.00175202 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96GW9, Q76E79, Q8IW62, Q8N7N4 | Gene names | MARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
SYM_HUMAN
|
||||||
| NC score | 0.151583 (rank : 15) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
MCA3_MOUSE
|
||||||
| NC score | 0.135944 (rank : 16) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D1M4 | Gene names | Eef1e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
SYM_MOUSE
|
||||||
| NC score | 0.106060 (rank : 17) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
MCA3_HUMAN
|
||||||
| NC score | 0.083380 (rank : 18) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43324, Q5THS2 | Gene names | EEF1E1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
SYEP_MOUSE
|
||||||
| NC score | 0.079501 (rank : 19) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
GSTT2_HUMAN
|
||||||
| NC score | 0.066750 (rank : 20) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30712, O60665, Q9HD76 | Gene names | GSTT2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
SYEP_HUMAN
|
||||||
| NC score | 0.049778 (rank : 21) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.023716 (rank : 22) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
ZAR1_HUMAN
|
||||||
| NC score | 0.019912 (rank : 23) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
IBP5_MOUSE
|
||||||
| NC score | 0.018224 (rank : 24) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.015914 (rank : 25) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.015905 (rank : 26) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.014540 (rank : 27) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.012849 (rank : 28) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
UBP14_HUMAN
|
||||||
| NC score | 0.011711 (rank : 29) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
DPOD3_HUMAN
|
||||||
| NC score | 0.011501 (rank : 30) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
TRPS1_HUMAN
|
||||||
| NC score | 0.011417 (rank : 31) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
CNTP1_MOUSE
|
||||||
| NC score | 0.010835 (rank : 32) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
PGTA_MOUSE
|
||||||
| NC score | 0.010607 (rank : 33) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHK4, Q9JLX2 | Gene names | Rabggta | |||
|
Domain Architecture |

|
|||||
| Description | Geranylgeranyl transferase type-2 alpha subunit (EC 2.5.1.60) (Geranylgeranyl transferase type II alpha subunit) (Rab geranylgeranyltransferase alpha subunit) (Rab geranyl- geranyltransferase alpha subunit) (Rab GG transferase alpha) (Rab GGTase alpha). | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.009355 (rank : 34) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
CNTP1_HUMAN
|
||||||
| NC score | 0.009068 (rank : 35) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.008407 (rank : 36) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.008348 (rank : 37) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
CN092_HUMAN
|
||||||
| NC score | 0.007767 (rank : 38) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.007533 (rank : 39) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.006677 (rank : 40) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
DYH9_HUMAN
|
||||||
| NC score | 0.006159 (rank : 41) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.002622 (rank : 42) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
ACM3_MOUSE
|
||||||
| NC score | 0.002276 (rank : 43) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERZ3, Q64055 | Gene names | Chrm3, Chrm-3 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M3 (Mm3 mAChR). | |||||
|
KS6B1_MOUSE
|
||||||
| NC score | 0.002180 (rank : 44) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BSK8, Q8CHX0 | Gene names | Rps6kb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase beta-1 (EC 2.7.11.1) (Ribosomal protein S6 kinase I) (S6K) (S6K1) (70 kDa ribosomal protein S6 kinase 1) (p70 S6 kinase alpha) (p70(S6K)-alpha) (p70-S6K) (P70S6K). | |||||
|
KS6B1_HUMAN
|
||||||
| NC score | 0.002179 (rank : 45) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23443 | Gene names | RPS6KB1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-1 (EC 2.7.11.1) (Ribosomal protein S6 kinase I) (S6K) (S6K1) (70 kDa ribosomal protein S6 kinase 1) (p70 S6 kinase alpha) (p70(S6K)-alpha) (p70-S6K) (P70S6K) (p70-alpha). | |||||
|
KS6B2_HUMAN
|
||||||
| NC score | 0.001636 (rank : 46) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBS0, O94809, Q9UEC1 | Gene names | RPS6KB2, STK14B | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (p70 S6 kinase beta) (S6K-beta) (p70-beta) (S6 kinase-related kinase) (SRK) (Serine/threonine-protein kinase 14 beta). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.001171 (rank : 47) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ZN232_HUMAN
|
||||||
| NC score | -0.000673 (rank : 48) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNY5 | Gene names | ZNF232, ZSCAN11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 232 (Zinc finger and SCAN domain-containing protein 11). | |||||