Please be patient as the page loads
|
H12_MOUSE
|
||||||
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
H12_MOUSE
|
||||||
| θ value | 6.17384e-33 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
H12_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 2) | NC score | 0.986930 (rank : 2) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
H14_MOUSE
|
||||||
| θ value | 4.42448e-31 (rank : 3) | NC score | 0.974254 (rank : 6) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
H13_HUMAN
|
||||||
| θ value | 7.54701e-31 (rank : 4) | NC score | 0.984394 (rank : 3) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
H13_MOUSE
|
||||||
| θ value | 7.54701e-31 (rank : 5) | NC score | 0.979684 (rank : 4) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
H14_HUMAN
|
||||||
| θ value | 7.54701e-31 (rank : 6) | NC score | 0.968592 (rank : 9) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
H15_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 7) | NC score | 0.969861 (rank : 8) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
H15_MOUSE
|
||||||
| θ value | 1.08979e-29 (rank : 8) | NC score | 0.963142 (rank : 10) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
H11_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 9) | NC score | 0.971197 (rank : 7) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
H11_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 10) | NC score | 0.978000 (rank : 5) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1. | |||||
|
H1T_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 11) | NC score | 0.956165 (rank : 12) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22492, Q8IUE8 | Gene names | HIST1H1T, H1FT, H1T | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
H1T_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 12) | NC score | 0.957810 (rank : 11) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q07133, Q8CGP8 | Gene names | Hist1h1t, H1ft, H1t | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
H10_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 13) | NC score | 0.819472 (rank : 14) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07305 | Gene names | H1F0, H1FV | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.0 (H1(0)) (Histone H1'). | |||||
|
H10_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 14) | NC score | 0.820683 (rank : 13) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10922 | Gene names | H1f0, H1fv | |||
|
Domain Architecture |
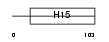
|
|||||
| Description | Histone H1' (H1.0) (H1(0)). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 15) | NC score | 0.134602 (rank : 18) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
H1X_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 16) | NC score | 0.805291 (rank : 15) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92522 | Gene names | H1FX | |||
|
Domain Architecture |
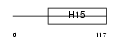
|
|||||
| Description | Histone H1x. | |||||
|
HILS1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 17) | NC score | 0.675101 (rank : 16) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYL0 | Gene names | Hils1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatid-specific linker histone H1-like protein (TISP64). | |||||
|
HILS1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 18) | NC score | 0.591938 (rank : 17) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P60008 | Gene names | HILS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatid-specific linker histone H1-like protein. | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 19) | NC score | 0.115046 (rank : 22) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 20) | NC score | 0.104447 (rank : 23) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 21) | NC score | 0.124828 (rank : 19) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.035799 (rank : 65) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
DPOD3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.100320 (rank : 25) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EQ28 | Gene names | Pold3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.119527 (rank : 20) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.019967 (rank : 70) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
FOXC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.020106 (rank : 69) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |
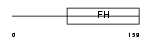
|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.086293 (rank : 31) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.091855 (rank : 29) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
IF16_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.062751 (rank : 44) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.070305 (rank : 40) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.097035 (rank : 27) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.040323 (rank : 64) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.044755 (rank : 61) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.068180 (rank : 41) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SUHW4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.023352 (rank : 67) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
CF150_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.089671 (rank : 30) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N884, Q32NC9, Q5SWL0, Q5SWL1, Q96E45 | Gene names | C6orf150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf150. | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.046035 (rank : 60) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.022395 (rank : 68) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.098556 (rank : 26) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.051656 (rank : 54) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NFM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.049032 (rank : 58) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.048508 (rank : 59) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SETD8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.032797 (rank : 66) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.044303 (rank : 62) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.012850 (rank : 72) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CD2L5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.004065 (rank : 73) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.119427 (rank : 21) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MO4L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.058767 (rank : 47) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.058006 (rank : 49) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
LYAR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.041917 (rank : 63) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.013070 (rank : 71) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
SYEP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.073041 (rank : 37) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
AFF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.054649 (rank : 50) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.080248 (rank : 33) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.076439 (rank : 35) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.073113 (rank : 36) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.050711 (rank : 56) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
DPOD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.050364 (rank : 57) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
GA2L3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.051217 (rank : 55) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
K1210_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.072385 (rank : 38) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
LAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.103375 (rank : 24) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.085558 (rank : 32) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.068127 (rank : 42) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.054405 (rank : 51) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.072186 (rank : 39) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.062984 (rank : 43) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.052722 (rank : 52) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.094005 (rank : 28) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.059481 (rank : 46) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.062466 (rank : 45) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.078648 (rank : 34) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.052044 (rank : 53) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
ZN638_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.058041 (rank : 48) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
H12_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.17384e-33 (rank : 1) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
H12_HUMAN
|
||||||
| NC score | 0.986930 (rank : 2) | θ value | 4.42448e-31 (rank : 2) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
H13_HUMAN
|
||||||
| NC score | 0.984394 (rank : 3) | θ value | 7.54701e-31 (rank : 4) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
H13_MOUSE
|
||||||
| NC score | 0.979684 (rank : 4) | θ value | 7.54701e-31 (rank : 5) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
H11_HUMAN
|
||||||
| NC score | 0.978000 (rank : 5) | θ value | 2.35151e-24 (rank : 10) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1. | |||||
|
H14_MOUSE
|
||||||
| NC score | 0.974254 (rank : 6) | θ value | 4.42448e-31 (rank : 3) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
H11_MOUSE
|
||||||
| NC score | 0.971197 (rank : 7) | θ value | 1.05554e-24 (rank : 9) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
H15_HUMAN
|
||||||
| NC score | 0.969861 (rank : 8) | θ value | 8.3442e-30 (rank : 7) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
H14_HUMAN
|
||||||
| NC score | 0.968592 (rank : 9) | θ value | 7.54701e-31 (rank : 6) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
H15_MOUSE
|
||||||
| NC score | 0.963142 (rank : 10) | θ value | 1.08979e-29 (rank : 8) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
H1T_MOUSE
|
||||||
| NC score | 0.957810 (rank : 11) | θ value | 3.75424e-22 (rank : 12) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q07133, Q8CGP8 | Gene names | Hist1h1t, H1ft, H1t | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
H1T_HUMAN
|
||||||
| NC score | 0.956165 (rank : 12) | θ value | 4.43474e-23 (rank : 11) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22492, Q8IUE8 | Gene names | HIST1H1T, H1FT, H1T | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
H10_MOUSE
|
||||||
| NC score | 0.820683 (rank : 13) | θ value | 4.76016e-09 (rank : 14) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10922 | Gene names | H1f0, H1fv | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1' (H1.0) (H1(0)). | |||||
|
H10_HUMAN
|
||||||
| NC score | 0.819472 (rank : 14) | θ value | 4.76016e-09 (rank : 13) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07305 | Gene names | H1F0, H1FV | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.0 (H1(0)) (Histone H1'). | |||||
|
H1X_HUMAN
|
||||||
| NC score | 0.805291 (rank : 15) | θ value | 4.45536e-07 (rank : 16) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92522 | Gene names | H1FX | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1x. | |||||
|
HILS1_MOUSE
|
||||||
| NC score | 0.675101 (rank : 16) | θ value | 9.92553e-07 (rank : 17) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYL0 | Gene names | Hils1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatid-specific linker histone H1-like protein (TISP64). | |||||
|
HILS1_HUMAN
|
||||||
| NC score | 0.591938 (rank : 17) | θ value | 1.43324e-05 (rank : 18) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P60008 | Gene names | HILS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatid-specific linker histone H1-like protein. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.134602 (rank : 18) | θ value | 1.80886e-08 (rank : 15) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.124828 (rank : 19) | θ value | 0.0113563 (rank : 21) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.119527 (rank : 20) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.119427 (rank : 21) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.115046 (rank : 22) | θ value | 0.00390308 (rank : 19) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.104447 (rank : 23) | θ value | 0.00390308 (rank : 20) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.103375 (rank : 24) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
DPOD3_MOUSE
|
||||||
| NC score | 0.100320 (rank : 25) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EQ28 | Gene names | Pold3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.098556 (rank : 26) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.097035 (rank : 27) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.094005 (rank : 28) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.091855 (rank : 29) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
CF150_HUMAN
|
||||||
| NC score | 0.089671 (rank : 30) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N884, Q32NC9, Q5SWL0, Q5SWL1, Q96E45 | Gene names | C6orf150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf150. | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.086293 (rank : 31) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.085558 (rank : 32) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.080248 (rank : 33) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.078648 (rank : 34) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.076439 (rank : 35) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.073113 (rank : 36) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
SYEP_MOUSE
|
||||||
| NC score | 0.073041 (rank : 37) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.072385 (rank : 38) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.072186 (rank : 39) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.070305 (rank : 40) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.068180 (rank : 41) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.068127 (rank : 42) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.062984 (rank : 43) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.062751 (rank : 44) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.062466 (rank : 45) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.059481 (rank : 46) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
MO4L2_MOUSE
|
||||||
| NC score | 0.058767 (rank : 47) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.058041 (rank : 48) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.058006 (rank : 49) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.054649 (rank : 50) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.054405 (rank : 51) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.052722 (rank : 52) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.052044 (rank : 53) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.051656 (rank : 54) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
GA2L3_HUMAN
|
||||||
| NC score | 0.051217 (rank : 55) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.050711 (rank : 56) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
DPOD3_HUMAN
|
||||||
| NC score | 0.050364 (rank : 57) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.049032 (rank : 58) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.048508 (rank : 59) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.046035 (rank : 60) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.044755 (rank : 61) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.044303 (rank : 62) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
LYAR_MOUSE
|
||||||
| NC score | 0.041917 (rank : 63) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.040323 (rank : 64) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.035799 (rank : 65) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
SETD8_HUMAN
|
||||||
| NC score | 0.032797 (rank : 66) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
SUHW4_HUMAN
|
||||||
| NC score | 0.023352 (rank : 67) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.022395 (rank : 68) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
FOXC2_HUMAN
|
||||||
| NC score | 0.020106 (rank : 69) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.019967 (rank : 70) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.013070 (rank : 71) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.012850 (rank : 72) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CD2L5_MOUSE
|
||||||
| NC score | 0.004065 (rank : 73) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||