Please be patient as the page loads
|
CF150_HUMAN
|
||||||
| SwissProt Accessions | Q8N884, Q32NC9, Q5SWL0, Q5SWL1, Q96E45 | Gene names | C6orf150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf150. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CF150_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q8N884, Q32NC9, Q5SWL0, Q5SWL1, Q96E45 | Gene names | C6orf150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf150. | |||||
|
CC059_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 2) | NC score | 0.284184 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYB1, Q86VD8 | Gene names | C3orf59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf59. | |||||
|
CC059_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 3) | NC score | 0.284580 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C525, Q5U4H4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf59 homolog. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 4) | NC score | 0.091649 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 5) | NC score | 0.068069 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.164854 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 7) | NC score | 0.052184 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.141010 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
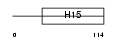
NFH_MOUSE
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.079703 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
H14_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.116362 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.037853 (rank : 118) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.102430 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.021658 (rank : 133) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
FBXL6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.065430 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N531, Q9H5W9, Q9UKC7 | Gene names | FBXL6, FBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 6 (F-box and leucine-rich repeat protein 6) (F-box protein FBL6) (FBL6A). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.106429 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.093387 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.041803 (rank : 113) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CSF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.081684 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.043271 (rank : 110) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.041166 (rank : 114) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.068305 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.081848 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AP2A2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.034832 (rank : 122) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.034163 (rank : 123) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CLIC6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.042986 (rank : 111) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.110767 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
NCA11_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.038511 (rank : 116) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
PRC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.053647 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43663, Q9BSB6 | Gene names | PRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
CARD6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.075561 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |
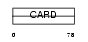
|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
H14_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.107240 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.042442 (rank : 112) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
RUSC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.050651 (rank : 103) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.019161 (rank : 138) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.047810 (rank : 106) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.081527 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
ARTN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.058529 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T4W7, O95441, O96030, Q6P6A3 | Gene names | ARTN, EVN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemin precursor (Enovin) (Neublastin). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.029264 (rank : 126) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.057629 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.096976 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
FOXI2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.015763 (rank : 142) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3I5G5, Q8BIK9 | Gene names | Foxi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein I2. | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.054142 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.087985 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.063426 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
U383_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.047774 (rank : 107) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYL2 | Gene names | C4orf23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative methyltransferase UPF0383 (EC 2.1.1.-). | |||||
|
ASPC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.044289 (rank : 109) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZE9, Q7Z6N7, Q8WV59, Q96LS5, Q96M40 | Gene names | ASPSCR1, ASPL, RCC17, TUG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tether containing UBX domain for GLUT4 (Alveolar soft part sarcoma chromosome region candidate 1) (Alveolar soft part sarcoma locus) (Renal papillary cell carcinoma protein 17). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.046964 (rank : 108) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.019887 (rank : 137) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
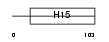
H15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.098435 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.065316 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.082094 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.062829 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
ZAR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.064878 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.078045 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.025937 (rank : 131) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
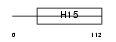
H12_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.089671 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |
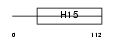
|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
H13_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.096800 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
MILK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.054698 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.081955 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SGIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.067312 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.100153 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.082522 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
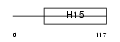
H15_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.092960 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.018041 (rank : 141) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.075608 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
ZCHC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.051479 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NUD5, Q6NT79 | Gene names | ZCCHC3, C20orf99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 3. | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.056575 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
BHLH4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.020780 (rank : 135) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
CK024_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.053366 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
EF1D_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.028682 (rank : 128) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57776, Q9CWW2, Q9CWY1, Q9CYD4, Q9CYJ5 | Gene names | Eef1d | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-delta (EF-1-delta). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.036534 (rank : 121) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RPOM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.029129 (rank : 127) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00411, O60370 | Gene names | POLRMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase, mitochondrial precursor (EC 2.7.7.6) (MtRPOL). | |||||
|
TCF17_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.000503 (rank : 149) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61751 | Gene names | Znf354a, Kid1, Tcf17, Zfp354a | |||
|
Domain Architecture |
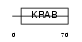
|
|||||
| Description | Zinc finger protein 354A (Transcription factor 17) (Renal transcription factor Kid-1) (Kidney, ischemia, and developmentally- regulated protein 1). | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.003903 (rank : 147) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.008873 (rank : 145) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.026466 (rank : 130) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
DBPA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.027714 (rank : 129) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JKB3, Q80WG4, Q9EQF7, Q9EQF8 | Gene names | Csda, Msy4, Ybx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding protein A (Cold shock domain-containing protein A) (Y-box protein 3). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.018495 (rank : 139) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
ICAL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.040605 (rank : 115) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
K1196_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.021139 (rank : 134) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
MAML1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.036537 (rank : 120) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.074767 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.052764 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RPGF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.020463 (rank : 136) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.038369 (rank : 117) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.077850 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
ADRM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.033720 (rank : 124) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JKV1 | Gene names | Adrm1, Gp110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110) (ARM-1). | |||||
|
APBA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.023203 (rank : 132) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.090002 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.037512 (rank : 119) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.002884 (rank : 148) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
GPVI_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.009849 (rank : 144) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCN6, Q9HCN7, Q9UIF2 | Gene names | GP6, GPVI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein VI precursor. | |||||
|
IF39_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.031778 (rank : 125) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.048486 (rank : 105) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
POF1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.013301 (rank : 143) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.018487 (rank : 140) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.008617 (rank : 146) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.052194 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.052783 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
BAG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.050986 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
BSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.070947 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.061146 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.058225 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.055114 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.050116 (rank : 104) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.054997 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
H10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.057354 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07305 | Gene names | H1F0, H1FV | |||
|
Domain Architecture |
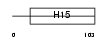
|
|||||
| Description | Histone H1.0 (H1(0)) (Histone H1'). | |||||
|
H10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.055605 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10922 | Gene names | H1f0, H1fv | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1' (H1.0) (H1(0)). | |||||
|
H11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.067247 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1. | |||||
|
H11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.068245 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
H12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.071832 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
H13_HUMAN
|
||||||
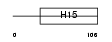
| θ value | θ > 10 (rank : 111) | NC score | 0.072778 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
H1T_HUMAN
|
||||||
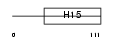
| θ value | θ > 10 (rank : 112) | NC score | 0.057142 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22492, Q8IUE8 | Gene names | HIST1H1T, H1FT, H1T | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
H1T_MOUSE
|
||||||
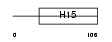
| θ value | θ > 10 (rank : 113) | NC score | 0.056651 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q07133, Q8CGP8 | Gene names | Hist1h1t, H1ft, H1t | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.053800 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.057573 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.059930 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.056384 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.068820 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.059126 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.055975 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.065921 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.061031 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MILK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.050975 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.082003 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.064393 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.063577 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.071978 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.053087 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.060734 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.059932 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.075660 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.056695 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.050951 (rank : 102) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.054663 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SELPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.064500 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.071526 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.074008 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.074322 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SYN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.052122 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.052577 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.055398 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.052797 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TAU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.051712 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TAU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.055289 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.071259 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.060705 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.060005 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.064253 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.059792 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
CF150_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q8N884, Q32NC9, Q5SWL0, Q5SWL1, Q96E45 | Gene names | C6orf150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf150. | |||||
|
CC059_MOUSE
|
||||||
| NC score | 0.284580 (rank : 2) | θ value | 7.1131e-05 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C525, Q5U4H4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf59 homolog. | |||||
|
CC059_HUMAN
|
||||||
| NC score | 0.284184 (rank : 3) | θ value | 5.44631e-05 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYB1, Q86VD8 | Gene names | C3orf59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf59. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.164854 (rank : 4) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.141010 (rank : 5) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
H14_HUMAN
|
||||||
| NC score | 0.116362 (rank : 6) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.110767 (rank : 7) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
H14_MOUSE
|
||||||
| NC score | 0.107240 (rank : 8) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.106429 (rank : 9) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.102430 (rank : 10) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.100153 (rank : 11) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
H15_HUMAN
|
||||||
| NC score | 0.098435 (rank : 12) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.096976 (rank : 13) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
H13_MOUSE
|
||||||
| NC score | 0.096800 (rank : 14) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.093387 (rank : 15) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
H15_MOUSE
|
||||||
| NC score | 0.092960 (rank : 16) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.091649 (rank : 17) | θ value | 0.00228821 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.090002 (rank : 18) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
H12_MOUSE
|
||||||
| NC score | 0.089671 (rank : 19) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.087985 (rank : 20) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.082522 (rank : 21) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.082094 (rank : 22) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.082003 (rank : 23) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.081955 (rank : 24) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.081848 (rank : 25) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.081684 (rank : 26) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.081527 (rank : 27) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.079703 (rank : 28) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.078045 (rank : 29) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.077850 (rank : 30) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.075660 (rank : 31) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.075608 (rank : 32) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
CARD6_HUMAN
|
||||||
| NC score | 0.075561 (rank : 33) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.074767 (rank : 34) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.074322 (rank : 35) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.074008 (rank : 36) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
H13_HUMAN
|
||||||
| NC score | 0.072778 (rank : 37) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.071978 (rank : 38) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
H12_HUMAN
|
||||||
| NC score | 0.071832 (rank : 39) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.071526 (rank : 40) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.071259 (rank : 41) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.070947 (rank : 42) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.068820 (rank : 43) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.068305 (rank : 44) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
H11_MOUSE
|
||||||
| NC score | 0.068245 (rank : 45) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.068069 (rank : 46) | θ value | 0.0113563 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
SGIP1_HUMAN
|
||||||
| NC score | 0.067312 (rank : 47) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
H11_HUMAN
|
||||||
| NC score | 0.067247 (rank : 48) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.065921 (rank : 49) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
FBXL6_HUMAN
|
||||||
| NC score | 0.065430 (rank : 50) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N531, Q9H5W9, Q9UKC7 | Gene names | FBXL6, FBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 6 (F-box and leucine-rich repeat protein 6) (F-box protein FBL6) (FBL6A). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.065316 (rank : 51) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
ZAR1_HUMAN
|
||||||
| NC score | 0.064878 (rank : 52) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.064500 (rank : 53) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.064393 (rank : 54) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.064253 (rank : 55) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.063577 (rank : 56) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.063426 (rank : 57) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.062829 (rank : 58) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.061146 (rank : 59) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.061031 (rank : 60) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.060734 (rank : 61) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.060705 (rank : 62) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.060005 (rank : 63) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.059932 (rank : 64) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.059930 (rank : 65) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.059792 (rank : 66) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.059126 (rank : 67) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
ARTN_HUMAN
|
||||||
| NC score | 0.058529 (rank : 68) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T4W7, O95441, O96030, Q6P6A3 | Gene names | ARTN, EVN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemin precursor (Enovin) (Neublastin). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.058225 (rank : 69) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.057629 (rank : 70) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.057573 (rank : 71) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
H10_HUMAN
|
||||||
| NC score | 0.057354 (rank : 72) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07305 | Gene names | H1F0, H1FV | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.0 (H1(0)) (Histone H1'). | |||||
|
H1T_HUMAN
|
||||||
| NC score | 0.057142 (rank : 73) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22492, Q8IUE8 | Gene names | HIST1H1T, H1FT, H1T | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.056695 (rank : 74) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
H1T_MOUSE
|
||||||
| NC score | 0.056651 (rank : 75) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q07133, Q8CGP8 | Gene names | Hist1h1t, H1ft, H1t | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.056575 (rank : 76) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.056384 (rank : 77) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.055975 (rank : 78) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
H10_MOUSE
|
||||||
| NC score | 0.055605 (rank : 79) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10922 | Gene names | H1f0, H1fv | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1' (H1.0) (H1(0)). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.055398 (rank : 80) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.055289 (rank : 81) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.055114 (rank : 82) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.054997 (rank : 83) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.054698 (rank : 84) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.054663 (rank : 85) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.054142 (rank : 86) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.053800 (rank : 87) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
PRC1_HUMAN
|
||||||
| NC score | 0.053647 (rank : 88) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43663, Q9BSB6 | Gene names | PRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.053366 (rank : 89) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.053087 (rank : 90) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.052797 (rank : 91) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.052783 (rank : 92) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.052764 (rank : 93) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.052577 (rank : 94) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.052194 (rank : 95) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.052184 (rank : 96) | θ value | 0.125558 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.052122 (rank : 97) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.051712 (rank : 98) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ZCHC3_HUMAN
|
||||||
| NC score | 0.051479 (rank : 99) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NUD5, Q6NT79 | Gene names | ZCCHC3, C20orf99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 3. | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.050986 (rank : 100) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.050975 (rank : 101) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.050951 (rank : 102) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RUSC2_HUMAN
|
||||||
| NC score | 0.050651 (rank : 103) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.050116 (rank : 104) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.048486 (rank : 105) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.047810 (rank : 106) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
U383_HUMAN
|
||||||
| NC score | 0.047774 (rank : 107) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYL2 | Gene names | C4orf23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative methyltransferase UPF0383 (EC 2.1.1.-). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.046964 (rank : 108) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
ASPC1_HUMAN
|
||||||
| NC score | 0.044289 (rank : 109) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZE9, Q7Z6N7, Q8WV59, Q96LS5, Q96M40 | Gene names | ASPSCR1, ASPL, RCC17, TUG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tether containing UBX domain for GLUT4 (Alveolar soft part sarcoma chromosome region candidate 1) (Alveolar soft part sarcoma locus) (Renal papillary cell carcinoma protein 17). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.043271 (rank : 110) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
CLIC6_MOUSE
|
||||||
| NC score | 0.042986 (rank : 111) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.042442 (rank : 112) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.041803 (rank : 113) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.041166 (rank : 114) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
ICAL_HUMAN
|
||||||
| NC score | 0.040605 (rank : 115) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
NCA11_MOUSE
|
||||||
| NC score | 0.038511 (rank : 116) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.038369 (rank : 117) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.037853 (rank : 118) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.037512 (rank : 119) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
MAML1_HUMAN
|
||||||
| NC score | 0.036537 (rank : 120) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.036534 (rank : 121) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AP2A2_HUMAN
|
||||||
| NC score | 0.034832 (rank : 122) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.034163 (rank : 123) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ADRM1_MOUSE
|
||||||
| NC score | 0.033720 (rank : 124) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JKV1 | Gene names | Adrm1, Gp110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110) (ARM-1). | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.031778 (rank : 125) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.029264 (rank : 126) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
RPOM_HUMAN
|
||||||
| NC score | 0.029129 (rank : 127) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00411, O60370 | Gene names | POLRMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase, mitochondrial precursor (EC 2.7.7.6) (MtRPOL). | |||||
|
EF1D_MOUSE
|
||||||
| NC score | 0.028682 (rank : 128) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57776, Q9CWW2, Q9CWY1, Q9CYD4, Q9CYJ5 | Gene names | Eef1d | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-delta (EF-1-delta). | |||||
|
DBPA_MOUSE
|
||||||
| NC score | 0.027714 (rank : 129) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JKB3, Q80WG4, Q9EQF7, Q9EQF8 | Gene names | Csda, Msy4, Ybx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding protein A (Cold shock domain-containing protein A) (Y-box protein 3). | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.026466 (rank : 130) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.025937 (rank : 131) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.023203 (rank : 132) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.021658 (rank : 133) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
K1196_HUMAN
|
||||||
| NC score | 0.021139 (rank : 134) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
BHLH4_HUMAN
|
||||||
| NC score | 0.020780 (rank : 135) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
RPGF6_HUMAN
|
||||||
| NC score | 0.020463 (rank : 136) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.019887 (rank : 137) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.019161 (rank : 138) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.018495 (rank : 139) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.018487 (rank : 140) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.018041 (rank : 141) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
FOXI2_MOUSE
|
||||||
| NC score | 0.015763 (rank : 142) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3I5G5, Q8BIK9 | Gene names | Foxi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein I2. | |||||
|
POF1B_HUMAN
|
||||||
| NC score | 0.013301 (rank : 143) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
GPVI_HUMAN
|
||||||
| NC score | 0.009849 (rank : 144) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCN6, Q9HCN7, Q9UIF2 | Gene names | GP6, GPVI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein VI precursor. | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.008873 (rank : 145) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.008617 (rank : 146) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.003903 (rank : 147) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.002884 (rank : 148) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
TCF17_MOUSE
|
||||||
| NC score | 0.000503 (rank : 149) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61751 | Gene names | Znf354a, Kid1, Tcf17, Zfp354a | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 354A (Transcription factor 17) (Renal transcription factor Kid-1) (Kidney, ischemia, and developmentally- regulated protein 1). | |||||