Please be patient as the page loads
|
ZCHC3_HUMAN
|
||||||
| SwissProt Accessions | Q9NUD5, Q6NT79 | Gene names | ZCCHC3, C20orf99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 3. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZCHC3_HUMAN
|
||||||
| θ value | 1.24043e-158 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NUD5, Q6NT79 | Gene names | ZCCHC3, C20orf99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 3. | |||||
|
CNBP_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 2) | NC score | 0.460288 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62633, P20694, Q5QJR0, Q6PJI7, Q96NV3 | Gene names | CNBP, ZNF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
CNBP_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 3) | NC score | 0.451534 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
ZCH13_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 4) | NC score | 0.429051 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WW36 | Gene names | ZCCHC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 13. | |||||
|
ZCHC9_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 5) | NC score | 0.352659 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N567, Q9H027 | Gene names | ZCCHC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
ZCHC9_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 6) | NC score | 0.340149 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R1J3, Q921T6 | Gene names | Zcchc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
ZN696_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.024129 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H7X3 | Gene names | ZNF696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 696. | |||||
|
ZCHC7_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.257149 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.034189 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.023301 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
IRX5_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.038619 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKQ4, Q80WV4, Q9JLL5 | Gene names | Irx5, Irxb2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2). | |||||
|
LN28A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.119830 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H9Z2 | Gene names | LIN28, CSDD1, LIN28A, ZCCHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Zinc finger CCHC domain-containing protein 1). | |||||
|
FBN2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.014021 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.023054 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.031028 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.007171 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ZNF16_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.018292 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17020, Q96FG0 | Gene names | ZNF16, KOX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 16 (Zinc finger protein KOX9). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.020472 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.081825 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.086399 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.038381 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
ZN205_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.017310 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95201 | Gene names | ZNF205, ZNF210 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 205 (Zinc finger protein 210). | |||||
|
LN28A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.113777 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K3Y3, Q6NV62 | Gene names | Lin28, Lin28a, Tex17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Testis-expressed protein 17). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.031433 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
ZN677_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.017034 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86XU0 | Gene names | ZNF677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 677. | |||||
|
CF150_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.051479 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N884, Q32NC9, Q5SWL0, Q5SWL1, Q96E45 | Gene names | C6orf150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf150. | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.020847 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CRIP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.020353 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52943 | Gene names | CRIP2, CRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich protein 2 (CRP2) (Protein ESP1). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.034564 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.031232 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RBM4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.056464 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.056491 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
ZCRB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.065413 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TBF4, Q6PJX0 | Gene names | ZCRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (U11/U12 snRNP 31 kDa protein) (U11/U12-31K). | |||||
|
ZCRB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.067381 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CZ96, Q3TDL3 | Gene names | Zcrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (MADP-1). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.033202 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
ZFP13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.016519 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P10754, Q80WS0, Q8CDD0 | Gene names | Zfp13, Krox-8, Zfp-13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 13 (Zfp-13) (Krox-8 protein). | |||||
|
ZN497_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.016424 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZNH5, Q6ZTD2, Q9UIA8 | Gene names | ZNF497 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 497. | |||||
|
ZN514_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.017555 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96K75 | Gene names | ZNF514 | |||
|
Domain Architecture |
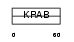
|
|||||
| Description | Zinc finger protein 514. | |||||
|
ZN672_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.017755 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q499Z4, Q96H65, Q96IM3, Q9H6G5 | Gene names | ZNF672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 672. | |||||
|
NUAK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.002613 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.017419 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
ULK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.003720 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
ZN365_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.023232 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BG89, Q80TQ4, Q8BK39, Q8BXT2 | Gene names | Znf365, Kiaa0844, Zfp365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365 (Su48). | |||||
|
ZN655_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.015584 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N720, Q8IV00, Q8TA89, Q96EZ3 | Gene names | ZNF655, VIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 655 (Vav-interacting Krueppel-like protein). | |||||
|
FXL19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.014896 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
GELS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.009641 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06396, Q8WVV7 | Gene names | GSN | |||
|
Domain Architecture |

|
|||||
| Description | Gelsolin precursor (Actin-depolymerizing factor) (ADF) (Brevin) (AGEL). | |||||
|
PRRT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.040860 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RBM4B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.052061 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.052022 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
SFRS7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.049284 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |
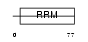
|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
SFRS7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.049688 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
ZN490_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.016000 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULM2 | Gene names | ZNF490, KIAA1198 | |||
|
Domain Architecture |
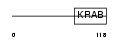
|
|||||
| Description | Zinc finger protein 490. | |||||
|
ZN679_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.014429 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYX0 | Gene names | ZNF679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 679. | |||||
|
LN28B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.078647 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZN17 | Gene names | LIN28B, CSDD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
LN28B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.077956 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q45KJ6, Q3UZC6, Q3V444 | Gene names | Lin28b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
ZCHC3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.24043e-158 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NUD5, Q6NT79 | Gene names | ZCCHC3, C20orf99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 3. | |||||
|
CNBP_HUMAN
|
||||||
| NC score | 0.460288 (rank : 2) | θ value | 3.41135e-07 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62633, P20694, Q5QJR0, Q6PJI7, Q96NV3 | Gene names | CNBP, ZNF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
CNBP_MOUSE
|
||||||
| NC score | 0.451534 (rank : 3) | θ value | 3.41135e-07 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
ZCH13_HUMAN
|
||||||
| NC score | 0.429051 (rank : 4) | θ value | 9.29e-05 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WW36 | Gene names | ZCCHC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 13. | |||||
|
ZCHC9_HUMAN
|
||||||
| NC score | 0.352659 (rank : 5) | θ value | 0.000786445 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N567, Q9H027 | Gene names | ZCCHC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
ZCHC9_MOUSE
|
||||||
| NC score | 0.340149 (rank : 6) | θ value | 0.00175202 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R1J3, Q921T6 | Gene names | Zcchc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
ZCHC7_HUMAN
|
||||||
| NC score | 0.257149 (rank : 7) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
LN28A_HUMAN
|
||||||
| NC score | 0.119830 (rank : 8) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H9Z2 | Gene names | LIN28, CSDD1, LIN28A, ZCCHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Zinc finger CCHC domain-containing protein 1). | |||||
|
LN28A_MOUSE
|
||||||
| NC score | 0.113777 (rank : 9) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K3Y3, Q6NV62 | Gene names | Lin28, Lin28a, Tex17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Testis-expressed protein 17). | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.086399 (rank : 10) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.081825 (rank : 11) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
LN28B_HUMAN
|
||||||
| NC score | 0.078647 (rank : 12) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZN17 | Gene names | LIN28B, CSDD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
LN28B_MOUSE
|
||||||
| NC score | 0.077956 (rank : 13) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q45KJ6, Q3UZC6, Q3V444 | Gene names | Lin28b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
ZCRB1_MOUSE
|
||||||
| NC score | 0.067381 (rank : 14) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CZ96, Q3TDL3 | Gene names | Zcrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (MADP-1). | |||||
|
ZCRB1_HUMAN
|
||||||
| NC score | 0.065413 (rank : 15) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TBF4, Q6PJX0 | Gene names | ZCRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (U11/U12 snRNP 31 kDa protein) (U11/U12-31K). | |||||
|
RBM4_MOUSE
|
||||||
| NC score | 0.056491 (rank : 16) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
RBM4_HUMAN
|
||||||
| NC score | 0.056464 (rank : 17) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4B_HUMAN
|
||||||
| NC score | 0.052061 (rank : 18) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| NC score | 0.052022 (rank : 19) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
CF150_HUMAN
|
||||||
| NC score | 0.051479 (rank : 20) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N884, Q32NC9, Q5SWL0, Q5SWL1, Q96E45 | Gene names | C6orf150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf150. | |||||
|
SFRS7_MOUSE
|
||||||
| NC score | 0.049688 (rank : 21) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
SFRS7_HUMAN
|
||||||
| NC score | 0.049284 (rank : 22) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
PRRT3_MOUSE
|
||||||
| NC score | 0.040860 (rank : 23) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
IRX5_MOUSE
|
||||||
| NC score | 0.038619 (rank : 24) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKQ4, Q80WV4, Q9JLL5 | Gene names | Irx5, Irxb2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.038381 (rank : 25) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.034564 (rank : 26) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.034189 (rank : 27) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.033202 (rank : 28) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.031433 (rank : 29) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.031232 (rank : 30) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.031028 (rank : 31) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
ZN696_HUMAN
|
||||||
| NC score | 0.024129 (rank : 32) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H7X3 | Gene names | ZNF696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 696. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.023301 (rank : 33) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ZN365_MOUSE
|
||||||
| NC score | 0.023232 (rank : 34) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BG89, Q80TQ4, Q8BK39, Q8BXT2 | Gene names | Znf365, Kiaa0844, Zfp365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365 (Su48). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.023054 (rank : 35) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.020847 (rank : 36) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.020472 (rank : 37) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CRIP2_HUMAN
|
||||||
| NC score | 0.020353 (rank : 38) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52943 | Gene names | CRIP2, CRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich protein 2 (CRP2) (Protein ESP1). | |||||
|
ZNF16_HUMAN
|
||||||
| NC score | 0.018292 (rank : 39) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17020, Q96FG0 | Gene names | ZNF16, KOX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 16 (Zinc finger protein KOX9). | |||||
|
ZN672_HUMAN
|
||||||
| NC score | 0.017755 (rank : 40) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q499Z4, Q96H65, Q96IM3, Q9H6G5 | Gene names | ZNF672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 672. | |||||
|
ZN514_HUMAN
|
||||||
| NC score | 0.017555 (rank : 41) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96K75 | Gene names | ZNF514 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 514. | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.017419 (rank : 42) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
ZN205_HUMAN
|
||||||
| NC score | 0.017310 (rank : 43) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95201 | Gene names | ZNF205, ZNF210 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 205 (Zinc finger protein 210). | |||||
|
ZN677_HUMAN
|
||||||
| NC score | 0.017034 (rank : 44) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86XU0 | Gene names | ZNF677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 677. | |||||
|
ZFP13_MOUSE
|
||||||
| NC score | 0.016519 (rank : 45) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P10754, Q80WS0, Q8CDD0 | Gene names | Zfp13, Krox-8, Zfp-13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 13 (Zfp-13) (Krox-8 protein). | |||||
|
ZN497_HUMAN
|
||||||
| NC score | 0.016424 (rank : 46) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZNH5, Q6ZTD2, Q9UIA8 | Gene names | ZNF497 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 497. | |||||
|
ZN490_HUMAN
|
||||||
| NC score | 0.016000 (rank : 47) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULM2 | Gene names | ZNF490, KIAA1198 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 490. | |||||
|
ZN655_HUMAN
|
||||||
| NC score | 0.015584 (rank : 48) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N720, Q8IV00, Q8TA89, Q96EZ3 | Gene names | ZNF655, VIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 655 (Vav-interacting Krueppel-like protein). | |||||
|
FXL19_MOUSE
|
||||||
| NC score | 0.014896 (rank : 49) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
ZN679_HUMAN
|
||||||
| NC score | 0.014429 (rank : 50) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYX0 | Gene names | ZNF679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 679. | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.014021 (rank : 51) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
GELS_HUMAN
|
||||||
| NC score | 0.009641 (rank : 52) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06396, Q8WVV7 | Gene names | GSN | |||
|
Domain Architecture |

|
|||||
| Description | Gelsolin precursor (Actin-depolymerizing factor) (ADF) (Brevin) (AGEL). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.007171 (rank : 53) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ULK1_HUMAN
|
||||||
| NC score | 0.003720 (rank : 54) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
NUAK1_HUMAN
|
||||||
| NC score | 0.002613 (rank : 55) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||