Please be patient as the page loads
|
RIOK1_HUMAN
|
||||||
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RIOK1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
RIOK3_HUMAN
|
||||||
| θ value | 1.96316e-71 (rank : 2) | NC score | 0.883734 (rank : 3) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14730, Q8IXN9 | Gene names | RIOK3, SUDD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO3 (EC 2.7.11.1) (RIO kinase 3) (sudD homolog). | |||||
|
RIOK3_MOUSE
|
||||||
| θ value | 4.37348e-71 (rank : 3) | NC score | 0.885837 (rank : 2) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBU3, Q8CIC1 | Gene names | Riok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO3 (EC 2.7.11.1) (RIO kinase 3). | |||||
|
RIOK2_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 4) | NC score | 0.420852 (rank : 4) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CQS5, Q91XF3 | Gene names | Riok2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
RIOK2_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 5) | NC score | 0.420495 (rank : 5) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BVS4, Q9NUT0 | Gene names | RIOK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 6) | NC score | 0.047508 (rank : 16) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
NUAK2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.017655 (rank : 68) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H093 | Gene names | NUAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1) (SNF1/AMP kinase-related kinase) (SNARK). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.035161 (rank : 28) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.043037 (rank : 19) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
RBM13_MOUSE
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.071954 (rank : 9) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGS0, Q921E0, Q9D5G6 | Gene names | Rbm13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAK16-like protein RBM13 (RNA-binding motif protein 13). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.049779 (rank : 11) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
NUAK2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.016233 (rank : 72) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BZN4, Q80ZW3, Q8CIC0, Q9DBV0 | Gene names | Nuak2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1). | |||||
|
KS6A4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.014223 (rank : 76) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75676, O75585, Q53ES8 | Gene names | RPS6KA4, MSK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase alpha-4 (EC 2.7.11.1) (Nuclear mitogen- and stress-activated protein kinase 2) (90 kDa ribosomal protein S6 kinase 4) (Ribosomal protein kinase B) (RSKB). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.087063 (rank : 7) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.091476 (rank : 6) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.032558 (rank : 36) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.044640 (rank : 18) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.029492 (rank : 43) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.037325 (rank : 26) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
KS6A4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.013808 (rank : 79) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2B9, Q91X18 | Gene names | Rps6ka4, Msk2 | |||
|
Domain Architecture |
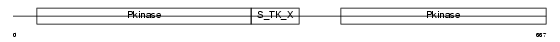
|
|||||
| Description | Ribosomal protein S6 kinase alpha-4 (EC 2.7.11.1) (Nuclear mitogen- and stress-activated protein kinase 2) (90 kDa ribosomal protein S6 kinase 4) (RSK-like protein kinase) (RLSK). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.026876 (rank : 52) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
NIM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.014343 (rank : 75) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IY84 | Gene names | NIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase NIM1 (EC 2.7.11.1). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.028995 (rank : 45) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.047395 (rank : 17) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.029800 (rank : 42) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.028713 (rank : 47) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
NIM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.015209 (rank : 73) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BHI9, Q8BXQ9 | Gene names | NIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase NIM1 (EC 2.7.11.1). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.028979 (rank : 46) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.029226 (rank : 44) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.052491 (rank : 10) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.041011 (rank : 24) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
SAPS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.035894 (rank : 27) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
G45IP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.048103 (rank : 14) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CR59, Q8BT05 | Gene names | Gadd45gip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth arrest and DNA-damage-inducible proteins-interacting protein 1. | |||||
|
IF38_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.048734 (rank : 12) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99613, O00215 | Gene names | EIF3S8 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110) (eIF3c). | |||||
|
IMUP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.081884 (rank : 8) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZP8, Q96H58, Q9HCR4 | Gene names | IMUP, C19orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immortalization-up-regulated protein (Hepatocyte growth factor activator inhibitor type 2-related small protein) (HAI-2-related small protein) (H2RSP). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.024302 (rank : 58) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CAPS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.032793 (rank : 34) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULU8, Q13339, Q6GQQ6, Q8N2Z5, Q8N3M7, Q8NFR0, Q96BC2 | Gene names | CADPS, CAPS, CAPS1, KIAA1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CAPS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.032771 (rank : 35) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TJ1, Q3TSP2, Q61374, Q6AXB4, Q6PGF0 | Gene names | Cadps, Caps, Caps1, Kiaa1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.023932 (rank : 59) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.024946 (rank : 57) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.043027 (rank : 20) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.030855 (rank : 40) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RP9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.042470 (rank : 22) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97762 | Gene names | Rp9, Rp9h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinitis pigmentosa 9 protein homolog (Pim-1-associated protein) (PAP-1). | |||||
|
SAPS3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.033781 (rank : 29) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
BCN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.033231 (rank : 33) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14457, O75595, Q9UNA8 | Gene names | BECN1, GT197 | |||
|
Domain Architecture |

|
|||||
| Description | Beclin-1 (Coiled-coil myosin-like BCL2-interacting protein) (Protein GT197). | |||||
|
CCD39_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.027138 (rank : 51) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
NFL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.017205 (rank : 69) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.030869 (rank : 39) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.026000 (rank : 54) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
TM1L1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.021918 (rank : 63) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.019507 (rank : 65) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
ZADH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.025997 (rank : 55) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8N7, Q6MZH8 | Gene names | ZADH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.027248 (rank : 50) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.033738 (rank : 30) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.023478 (rank : 60) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.033362 (rank : 32) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
FYB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.026857 (rank : 53) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.020721 (rank : 64) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
IPK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.037527 (rank : 25) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BMY7 | Gene names | Spink2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor. | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.016235 (rank : 71) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.032356 (rank : 37) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.047731 (rank : 15) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.042726 (rank : 21) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.013920 (rank : 78) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.014757 (rank : 74) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
NFL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.016685 (rank : 70) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.018219 (rank : 67) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.008750 (rank : 86) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.041738 (rank : 23) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.022137 (rank : 62) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.030035 (rank : 41) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.048381 (rank : 13) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.027417 (rank : 49) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.023341 (rank : 61) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TRI16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.011140 (rank : 81) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95361, Q7Z6I2, Q96BE8, Q96J43 | Gene names | TRIM16, EBBP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
TTC14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.025203 (rank : 56) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
BCN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.027547 (rank : 48) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88597 | Gene names | Becn1 | |||
|
Domain Architecture |

|
|||||
| Description | Beclin-1 (Coiled-coil myosin-like BCL2-interacting protein). | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.010859 (rank : 82) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.010105 (rank : 83) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.033678 (rank : 31) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.009146 (rank : 85) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.013059 (rank : 80) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.009398 (rank : 84) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.030949 (rank : 38) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.019098 (rank : 66) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
SUGT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.013988 (rank : 77) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |
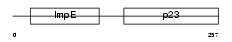
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
RIOK1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
RIOK3_MOUSE
|
||||||
| NC score | 0.885837 (rank : 2) | θ value | 4.37348e-71 (rank : 3) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBU3, Q8CIC1 | Gene names | Riok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO3 (EC 2.7.11.1) (RIO kinase 3). | |||||
|
RIOK3_HUMAN
|
||||||
| NC score | 0.883734 (rank : 3) | θ value | 1.96316e-71 (rank : 2) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14730, Q8IXN9 | Gene names | RIOK3, SUDD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO3 (EC 2.7.11.1) (RIO kinase 3) (sudD homolog). | |||||
|
RIOK2_MOUSE
|
||||||
| NC score | 0.420852 (rank : 4) | θ value | 3.29651e-10 (rank : 4) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CQS5, Q91XF3 | Gene names | Riok2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
RIOK2_HUMAN
|
||||||
| NC score | 0.420495 (rank : 5) | θ value | 9.59137e-10 (rank : 5) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BVS4, Q9NUT0 | Gene names | RIOK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.091476 (rank : 6) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.087063 (rank : 7) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
IMUP_HUMAN
|
||||||
| NC score | 0.081884 (rank : 8) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZP8, Q96H58, Q9HCR4 | Gene names | IMUP, C19orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immortalization-up-regulated protein (Hepatocyte growth factor activator inhibitor type 2-related small protein) (HAI-2-related small protein) (H2RSP). | |||||
|
RBM13_MOUSE
|
||||||
| NC score | 0.071954 (rank : 9) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGS0, Q921E0, Q9D5G6 | Gene names | Rbm13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAK16-like protein RBM13 (RNA-binding motif protein 13). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.052491 (rank : 10) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.049779 (rank : 11) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
IF38_HUMAN
|
||||||
| NC score | 0.048734 (rank : 12) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99613, O00215 | Gene names | EIF3S8 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110) (eIF3c). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.048381 (rank : 13) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
G45IP_MOUSE
|
||||||
| NC score | 0.048103 (rank : 14) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CR59, Q8BT05 | Gene names | Gadd45gip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth arrest and DNA-damage-inducible proteins-interacting protein 1. | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.047731 (rank : 15) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.047508 (rank : 16) | θ value | 0.0113563 (rank : 6) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.047395 (rank : 17) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.044640 (rank : 18) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.043037 (rank : 19) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.043027 (rank : 20) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.042726 (rank : 21) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
RP9_MOUSE
|
||||||
| NC score | 0.042470 (rank : 22) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97762 | Gene names | Rp9, Rp9h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinitis pigmentosa 9 protein homolog (Pim-1-associated protein) (PAP-1). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.041738 (rank : 23) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.041011 (rank : 24) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
IPK2_MOUSE
|
||||||
| NC score | 0.037527 (rank : 25) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BMY7 | Gene names | Spink2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.037325 (rank : 26) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
SAPS3_HUMAN
|
||||||
| NC score | 0.035894 (rank : 27) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.035161 (rank : 28) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
SAPS3_MOUSE
|
||||||
| NC score | 0.033781 (rank : 29) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.033738 (rank : 30) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.033678 (rank : 31) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.033362 (rank : 32) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
BCN1_HUMAN
|
||||||
| NC score | 0.033231 (rank : 33) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14457, O75595, Q9UNA8 | Gene names | BECN1, GT197 | |||
|
Domain Architecture |

|
|||||
| Description | Beclin-1 (Coiled-coil myosin-like BCL2-interacting protein) (Protein GT197). | |||||
|
CAPS1_HUMAN
|
||||||
| NC score | 0.032793 (rank : 34) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULU8, Q13339, Q6GQQ6, Q8N2Z5, Q8N3M7, Q8NFR0, Q96BC2 | Gene names | CADPS, CAPS, CAPS1, KIAA1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CAPS1_MOUSE
|
||||||
| NC score | 0.032771 (rank : 35) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TJ1, Q3TSP2, Q61374, Q6AXB4, Q6PGF0 | Gene names | Cadps, Caps, Caps1, Kiaa1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.032558 (rank : 36) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.032356 (rank : 37) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.030949 (rank : 38) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.030869 (rank : 39) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.030855 (rank : 40) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.030035 (rank : 41) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.029800 (rank : 42) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.029492 (rank : 43) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.029226 (rank : 44) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.028995 (rank : 45) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.028979 (rank : 46) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.028713 (rank : 47) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
BCN1_MOUSE
|
||||||
| NC score | 0.027547 (rank : 48) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88597 | Gene names | Becn1 | |||
|
Domain Architecture |

|
|||||
| Description | Beclin-1 (Coiled-coil myosin-like BCL2-interacting protein). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.027417 (rank : 49) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.027248 (rank : 50) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.027138 (rank : 51) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.026876 (rank : 52) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.026857 (rank : 53) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.026000 (rank : 54) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
ZADH1_HUMAN
|
||||||
| NC score | 0.025997 (rank : 55) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8N7, Q6MZH8 | Gene names | ZADH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.025203 (rank : 56) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.024946 (rank : 57) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.024302 (rank : 58) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.023932 (rank : 59) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.023478 (rank : 60) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.023341 (rank : 61) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.022137 (rank : 62) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TM1L1_MOUSE
|
||||||
| NC score | 0.021918 (rank : 63) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.020721 (rank : 64) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.019507 (rank : 65) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.019098 (rank : 66) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.018219 (rank : 67) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NUAK2_HUMAN
|
||||||
| NC score | 0.017655 (rank : 68) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H093 | Gene names | NUAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1) (SNF1/AMP kinase-related kinase) (SNARK). | |||||
|
NFL_MOUSE
|
||||||
| NC score | 0.017205 (rank : 69) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
NFL_HUMAN
|
||||||
| NC score | 0.016685 (rank : 70) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.016235 (rank : 71) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NUAK2_MOUSE
|
||||||
| NC score | 0.016233 (rank : 72) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BZN4, Q80ZW3, Q8CIC0, Q9DBV0 | Gene names | Nuak2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1). | |||||
|
NIM1_MOUSE
|
||||||
| NC score | 0.015209 (rank : 73) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BHI9, Q8BXQ9 | Gene names | NIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase NIM1 (EC 2.7.11.1). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.014757 (rank : 74) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
NIM1_HUMAN
|
||||||
| NC score | 0.014343 (rank : 75) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IY84 | Gene names | NIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase NIM1 (EC 2.7.11.1). | |||||
|
KS6A4_HUMAN
|
||||||
| NC score | 0.014223 (rank : 76) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75676, O75585, Q53ES8 | Gene names | RPS6KA4, MSK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase alpha-4 (EC 2.7.11.1) (Nuclear mitogen- and stress-activated protein kinase 2) (90 kDa ribosomal protein S6 kinase 4) (Ribosomal protein kinase B) (RSKB). | |||||
|
SUGT1_MOUSE
|
||||||
| NC score | 0.013988 (rank : 77) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.013920 (rank : 78) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
KS6A4_MOUSE
|
||||||
| NC score | 0.013808 (rank : 79) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2B9, Q91X18 | Gene names | Rps6ka4, Msk2 | |||
|
Domain Architecture |
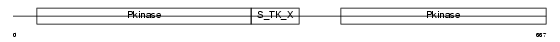
|
|||||
| Description | Ribosomal protein S6 kinase alpha-4 (EC 2.7.11.1) (Nuclear mitogen- and stress-activated protein kinase 2) (90 kDa ribosomal protein S6 kinase 4) (RSK-like protein kinase) (RLSK). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.013059 (rank : 80) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
TRI16_HUMAN
|
||||||
| NC score | 0.011140 (rank : 81) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95361, Q7Z6I2, Q96BE8, Q96J43 | Gene names | TRIM16, EBBP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.010859 (rank : 82) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.010105 (rank : 83) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.009398 (rank : 84) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.009146 (rank : 85) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.008750 (rank : 86) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||