Please be patient as the page loads
|
RIOK2_HUMAN
|
||||||
| SwissProt Accessions | Q9BVS4, Q9NUT0 | Gene names | RIOK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RIOK2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q9BVS4, Q9NUT0 | Gene names | RIOK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
RIOK2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.959589 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CQS5, Q91XF3 | Gene names | Riok2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
RIOK1_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 3) | NC score | 0.420495 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
RIOK3_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 4) | NC score | 0.352499 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBU3, Q8CIC1 | Gene names | Riok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO3 (EC 2.7.11.1) (RIO kinase 3). | |||||
|
RIOK3_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 5) | NC score | 0.343546 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14730, Q8IXN9 | Gene names | RIOK3, SUDD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO3 (EC 2.7.11.1) (RIO kinase 3) (sudD homolog). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 6) | NC score | 0.144803 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
NRK_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 7) | NC score | 0.071649 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 8) | NC score | 0.121501 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
NRK_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 9) | NC score | 0.069207 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9R0G8, Q6NV55, Q8C9S9, Q9R0S4 | Gene names | Nrk, Nesk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1) (Nck-interacting kinase-like embryo specific kinase) (NIK-like embryo-specific kinase) (NESK). | |||||
|
BMS1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.107250 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14692, Q86XJ9 | Gene names | BMS1L, KIAA0187 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BMS1 homolog. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 11) | NC score | 0.064604 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.080032 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.073617 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.044256 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
M4K3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.058446 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IVH8, Q8IVH7, Q9UDM5, Q9Y6R5 | Gene names | MAP4K3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 3 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 3) (MEK kinase kinase 3) (MEKKK 3) (Germinal center kinase-related protein kinase) (GLK). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.045507 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CHK1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.050735 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O35280, O54925, Q8CI40, Q9D0N2 | Gene names | Chek1, Chk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Chk1 (EC 2.7.11.1). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.039307 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.044636 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.058318 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
TF2AY_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.052056 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4I4, Q99PM2, Q9DAF4 | Gene names | Gtf2a1lf, Alf | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.024580 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
KPCB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.055800 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P05771, O43744, P05127, Q15138, Q93060, Q9UE49, Q9UE50, Q9UEH8, Q9UJ30, Q9UJ33 | Gene names | PRKCB1, PKCB, PRKCB | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
KPCB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.055943 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P68404, P04410, P04411 | Gene names | Prkcb1, Pkcb, Prkcb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.033393 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
FBXW7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.012333 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.074942 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
KPCA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.055062 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P17252, Q15137, Q96RE4 | Gene names | PRKCA, PKCA, PRKACA | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.039348 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
TTK_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.045516 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P33981 | Gene names | TTK, MPS1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (Phosphotyrosine picked threonine-protein kinase) (PYT). | |||||
|
ASPX_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.048788 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50289, Q9DAM6 | Gene names | Acrv1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1) (MSA- 63). | |||||
|
EXOC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.046989 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D4H1 | Gene names | Exoc2, Sec5, Sec5l1 | |||
|
Domain Architecture |
|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
MASTL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.058362 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96GX5, Q5T8D5, Q5T8D7, Q8NCD6, Q96SJ5 | Gene names | MASTL, THC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
TNNT3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.033604 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
TSSK4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.047263 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D411 | Gene names | Tssk4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine/threonine-protein kinase 4 (EC 2.7.11.1) (TSSK- 4) (Testis-specific kinase 4) (TSK-4). | |||||
|
CO5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.024462 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06684 | Gene names | C5, Hc | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor (Hemolytic complement) [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
GP158_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.028201 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
HUNK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.046898 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O88866 | Gene names | Hunk, Makv | |||
|
Domain Architecture |
|
|||||
| Description | Hormonally up-regulated neu tumor-associated kinase (EC 2.7.11.1) (Serine/threonine-protein kinase MAK-V). | |||||
|
KPCE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.054415 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02156, Q53SL4, Q53SM5, Q9UE81 | Gene names | PRKCE, PKCE | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
KPCE_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.054167 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P16054 | Gene names | Prkce, Pkce, Pkcea | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
SCG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.039672 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.037116 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
E2AK4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.056358 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1012 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P2K8, Q69YL7, Q6DC97, Q96GN6, Q9H5K1, Q9NSQ3, Q9NSZ5, Q9UJ56 | Gene names | EIF2AK4, GCN2, KIAA1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein). | |||||
|
E2AK4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.057642 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QZ05, Q6GQT4, Q6ZPT5, Q8C5S0, Q8CIF5, Q9CT30, Q9CUV9, Q9ESB6, Q9ESB7, Q9ESB8 | Gene names | Eif2ak4, Gcn2, Kiaa1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein) (mGCN2). | |||||
|
HUNK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.046398 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P57058 | Gene names | HUNK, MAKV | |||
|
Domain Architecture |
|
|||||
| Description | Hormonally up-regulated neu tumor-associated kinase (EC 2.7.11.1) (Serine/threonine-protein kinase MAK-V) (B19). | |||||
|
KKCC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.050050 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96RR4, O94883, Q8IUG2, Q8IUG3, Q8N3I4, Q8WY03, Q8WY04, Q8WY05, Q8WY06, Q96RP1, Q96RP2, Q96RR3, Q9BWE9, Q9UER3, Q9UES2, Q9Y5N2 | Gene names | CAMKK2, CAMKKB, KIAA0787 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 2 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase beta) (CaM-kinase kinase beta) (CaM-KK beta) (CaMKK beta). | |||||
|
KKCC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.050087 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8C078, Q80TS0, Q8BXM8, Q8C0G3, Q8CH42, Q8QZT7 | Gene names | Camkk2, Kiaa0787 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 2 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase beta) (CaM-kinase kinase beta) (CaM-KK beta) (CaMKK beta). | |||||
|
M4K2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.054911 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61161 | Gene names | Map4k2, Rab8ip | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 2 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 2) (MEK kinase kinase 2) (MEKKK 2) (Germinal center kinase) (GCK) (Rab8-interacting protein). | |||||
|
MASTL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.057195 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8C0P0, Q5RJW0, Q6NXX9, Q8BVF3, Q9CZH9, Q9D9V0 | Gene names | Mastl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
SSRP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.031264 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.032311 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
TAI12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.031695 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.035390 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.035963 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.027824 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CCKAR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.001141 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08786, Q8VCC7, Q9DBV6 | Gene names | Cckar | |||
|
Domain Architecture |
|
|||||
| Description | Cholecystokinin type A receptor (CCK-A receptor) (CCK-AR) (Cholecystokinin-1 receptor) (CCK1-R). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.020977 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.045545 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14757 | Gene names | CHEK1, CHK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Chk1 (EC 2.7.11.1). | |||||
|
KS6B2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.048311 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z1M4 | Gene names | Rps6kb2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K-beta 2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (S6K2). | |||||
|
LAS1L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.038834 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4W2, Q5JXQ0, Q8TEN5, Q9H9V5 | Gene names | LAS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LAS1-like protein. | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.012209 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.022957 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.065043 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
ST32B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.049017 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NY57, Q6UXH3, Q8IY14 | Gene names | STK32B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 32B (EC 2.7.11.1) (YANK2). | |||||
|
ST32B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.048706 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JJX8, Q7TMD3, Q8C4E0 | Gene names | Stk32b, Stk32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 32B (EC 2.7.11.1). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.037359 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
EXOC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.040932 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KP1, Q5JPC8, Q96AN6, Q9NUZ8, Q9UJM7 | Gene names | EXOC2, SEC5, SEC5L1 | |||
|
Domain Architecture |
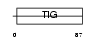
|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.013545 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.063410 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KPCA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.052900 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P20444 | Gene names | Prkca, Pkca | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
KPCD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.051620 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q05655, Q15144 | Gene names | PRKCD | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C delta type (EC 2.7.11.13) (nPKC-delta). | |||||
|
M4K5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.052652 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y4K4, Q8IYF6 | Gene names | MAP4K5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 5 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 5) (MEK kinase kinase 5) (MEKKK 5) (Kinase homologous to SPS1/STE20) (KHS). | |||||
|
M4K5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.052907 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BPM2, Q6PEQ2, Q8C3U5, Q8CGF3, Q99LM7, Q9CX73 | Gene names | Map4k5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 5 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 5) (MEK kinase kinase 5) (MEKKK 5). | |||||
|
MAVS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.025492 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.028661 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ST32A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.047497 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 834 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WU08 | Gene names | STK32A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 32A (EC 2.7.11.1) (YANK1). | |||||
|
STK39_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.045250 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |
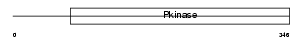
|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
TAI12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.029108 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGQ2, Q3UIH1 | Gene names | Taip12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.008764 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.042689 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.019640 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CJ078_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.026923 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86XK3, Q5JT40, Q8WW47 | Gene names | C10orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.014866 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
KPCG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.053161 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1126 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P05129 | Gene names | PRKCG, PKCG | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
KPCG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.053139 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P63318, P05697 | Gene names | Prkcg, Pkcc, Pkcg, Prkcc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
KPCT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.052363 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q04759, Q3MJF1, Q64FY5, Q9H508, Q9H549 | Gene names | PRKCQ, PRKCT | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.050185 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.049977 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.051177 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
TAOK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.043705 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1494 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9H2K8, Q658N1, Q8IUM4, Q9HC79, Q9NZM9, Q9UHG7 | Gene names | TAOK3, DPK, JIK, KDS, MAP3K18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3) (Jun kinase-inhibitory kinase) (JNK/SAPK- inhibitory kinase) (Dendritic cell-derived protein kinase) (Cutaneous T-cell lymphoma tumor antigen HD-CL-09) (CTCL tumor antigen HD-CL-09) (Kinase from chicken homolog A) (hKFC-A). | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.015918 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TNNT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.026654 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P45378, Q12975, Q12976, Q12977, Q12978, Q86TH6 | Gene names | TNNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT) (Beta TnTF). | |||||
|
AN32B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.013764 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
HNRPU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.014361 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.049519 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
MP2K7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.045063 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14733, O14648, O14816, O60452, O60453 | Gene names | MAP2K7, JNKK2, MKK7, PRKMK7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity mitogen-activated protein kinase kinase 7 (EC 2.7.12.2) (MAP kinase kinase 7) (MAPKK 7) (MAPK/ERK kinase 7) (JNK-activating kinase 2) (c-Jun N-terminal kinase kinase 2) (JNK kinase 2) (JNKK 2). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.023324 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.011669 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.023391 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TTK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.046166 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P35761 | Gene names | Ttk, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (ESK) (PYT). | |||||
|
VPS54_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.017216 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SPW0, Q8BPB3, Q8CFZ7, Q8CHL5, Q8R3R4, Q8R3X1 | Gene names | Vps54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 54 (Tumor antigen SLP-8p homolog). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.019284 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CDK7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.037670 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P50613, Q9BS60, Q9UE19 | Gene names | CDK7, MO15 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division protein kinase 7 (EC 2.7.11.22) (EC 2.7.11.23) (CDK- activating kinase) (CAK) (TFIIH basal transcription factor complex kinase subunit) (39 kDa protein kinase) (P39 Mo15) (STK1) (CAK1). | |||||
|
CDK7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.037834 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q03147, Q99KK3 | Gene names | Cdk7, Cdkn7, Crk4, Mo15, Mpk-7 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division protein kinase 7 (EC 2.7.11.22) (EC 2.7.11.23) (CDK- activating kinase) (CAK) (TFIIH basal transcription factor complex kinase subunit) (39 kDa protein kinase) (P39 Mo15) (Protein-tyrosine kinase MPK-7) (CR4 protein kinase) (CRK4). | |||||
|
CF153_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.025160 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96EU6, Q9BRF6, Q9P0C8 | Gene names | C6orf153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf153. | |||||
|
CF153_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.023993 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UFY0, Q91X71 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf153 homolog. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.015870 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HXC10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.004453 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P31257, P31312, Q7TMT7 | Gene names | Hoxc10, Hox-3.6, Hoxc-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein Hox-C10 (Hox-3.6). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.064116 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KPCD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.050967 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P28867, Q91V85, Q9Z333 | Gene names | Prkcd, Pkcd | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C delta type (EC 2.7.11.13) (nPKC-delta). | |||||
|
KPCT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.050693 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q02111 | Gene names | Prkcq, Pkcq | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||
|
LNP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.015566 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9C0E8, Q2M2V8, Q2YD99, Q658W8, Q8N5V9, Q96MS5 | Gene names | LNP, KIAA1715 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark. | |||||
|
M4K2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.052455 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q12851 | Gene names | MAP4K2, GCK, RAB8IP | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 2 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 2) (MEK kinase kinase 2) (MEKKK 2) (Germinal center kinase) (GC kinase) (Rab8-interacting protein) (B lymphocyte serine/threonine-protein kinase). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.048409 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.047991 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.019965 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.009124 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PAK7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.045445 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8C015, Q3TQJ7, Q6RWS7 | Gene names | Pak7, Pak5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (p21-activated kinase 5). | |||||
|
PLXB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.005109 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
PTPRG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.003355 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
RBM28_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.008477 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NW13, Q96CV3 | Gene names | RBM28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.012067 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.014919 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.014754 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
ST32A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.046388 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BGW6, Q7TPQ4 | Gene names | Stk32a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 32A (EC 2.7.11.1). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.055232 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.059636 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.059835 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RIOK2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q9BVS4, Q9NUT0 | Gene names | RIOK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
RIOK2_MOUSE
|
||||||
| NC score | 0.959589 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CQS5, Q91XF3 | Gene names | Riok2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
RIOK1_HUMAN
|
||||||
| NC score | 0.420495 (rank : 3) | θ value | 9.59137e-10 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
RIOK3_MOUSE
|
||||||
| NC score | 0.352499 (rank : 4) | θ value | 5.81887e-07 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBU3, Q8CIC1 | Gene names | Riok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO3 (EC 2.7.11.1) (RIO kinase 3). | |||||
|
RIOK3_HUMAN
|
||||||
| NC score | 0.343546 (rank : 5) | θ value | 2.21117e-06 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14730, Q8IXN9 | Gene names | RIOK3, SUDD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO3 (EC 2.7.11.1) (RIO kinase 3) (sudD homolog). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.144803 (rank : 6) | θ value | 3.19293e-05 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.121501 (rank : 7) | θ value | 0.00665767 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
BMS1_HUMAN
|
||||||
| NC score | 0.107250 (rank : 8) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14692, Q86XJ9 | Gene names | BMS1L, KIAA0187 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BMS1 homolog. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.080032 (rank : 9) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.074942 (rank : 10) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.073617 (rank : 11) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
NRK_HUMAN
|
||||||
| NC score | 0.071649 (rank : 12) | θ value | 0.00102713 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
NRK_MOUSE
|
||||||
| NC score | 0.069207 (rank : 13) | θ value | 0.0148317 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9R0G8, Q6NV55, Q8C9S9, Q9R0S4 | Gene names | Nrk, Nesk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1) (Nck-interacting kinase-like embryo specific kinase) (NIK-like embryo-specific kinase) (NESK). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.065043 (rank : 14) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.064604 (rank : 15) | θ value | 0.0252991 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.064116 (rank : 16) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.063410 (rank : 17) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.059835 (rank : 18) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.059636 (rank : 19) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
M4K3_HUMAN
|
||||||
| NC score | 0.058446 (rank : 20) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IVH8, Q8IVH7, Q9UDM5, Q9Y6R5 | Gene names | MAP4K3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 3 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 3) (MEK kinase kinase 3) (MEKKK 3) (Germinal center kinase-related protein kinase) (GLK). | |||||
|
MASTL_HUMAN
|
||||||
| NC score | 0.058362 (rank : 21) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96GX5, Q5T8D5, Q5T8D7, Q8NCD6, Q96SJ5 | Gene names | MASTL, THC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.058318 (rank : 22) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
E2AK4_MOUSE
|
||||||
| NC score | 0.057642 (rank : 23) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QZ05, Q6GQT4, Q6ZPT5, Q8C5S0, Q8CIF5, Q9CT30, Q9CUV9, Q9ESB6, Q9ESB7, Q9ESB8 | Gene names | Eif2ak4, Gcn2, Kiaa1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein) (mGCN2). | |||||
|
MASTL_MOUSE
|
||||||
| NC score | 0.057195 (rank : 24) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8C0P0, Q5RJW0, Q6NXX9, Q8BVF3, Q9CZH9, Q9D9V0 | Gene names | Mastl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
E2AK4_HUMAN
|
||||||
| NC score | 0.056358 (rank : 25) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1012 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P2K8, Q69YL7, Q6DC97, Q96GN6, Q9H5K1, Q9NSQ3, Q9NSZ5, Q9UJ56 | Gene names | EIF2AK4, GCN2, KIAA1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein). | |||||
|
KPCB_MOUSE
|
||||||
| NC score | 0.055943 (rank : 26) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P68404, P04410, P04411 | Gene names | Prkcb1, Pkcb, Prkcb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
KPCB_HUMAN
|
||||||
| NC score | 0.055800 (rank : 27) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P05771, O43744, P05127, Q15138, Q93060, Q9UE49, Q9UE50, Q9UEH8, Q9UJ30, Q9UJ33 | Gene names | PRKCB1, PKCB, PRKCB | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.055232 (rank : 28) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
KPCA_HUMAN
|
||||||
| NC score | 0.055062 (rank : 29) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P17252, Q15137, Q96RE4 | Gene names | PRKCA, PKCA, PRKACA | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
M4K2_MOUSE
|
||||||
| NC score | 0.054911 (rank : 30) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61161 | Gene names | Map4k2, Rab8ip | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 2 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 2) (MEK kinase kinase 2) (MEKKK 2) (Germinal center kinase) (GCK) (Rab8-interacting protein). | |||||
|
KPCE_HUMAN
|
||||||
| NC score | 0.054415 (rank : 31) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02156, Q53SL4, Q53SM5, Q9UE81 | Gene names | PRKCE, PKCE | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
KPCE_MOUSE
|
||||||
| NC score | 0.054167 (rank : 32) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P16054 | Gene names | Prkce, Pkce, Pkcea | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
KPCG_HUMAN
|
||||||
| NC score | 0.053161 (rank : 33) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1126 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P05129 | Gene names | PRKCG, PKCG | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
KPCG_MOUSE
|
||||||
| NC score | 0.053139 (rank : 34) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P63318, P05697 | Gene names | Prkcg, Pkcc, Pkcg, Prkcc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
M4K5_MOUSE
|
||||||
| NC score | 0.052907 (rank : 35) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BPM2, Q6PEQ2, Q8C3U5, Q8CGF3, Q99LM7, Q9CX73 | Gene names | Map4k5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 5 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 5) (MEK kinase kinase 5) (MEKKK 5). | |||||
|
KPCA_MOUSE
|
||||||
| NC score | 0.052900 (rank : 36) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P20444 | Gene names | Prkca, Pkca | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
M4K5_HUMAN
|
||||||
| NC score | 0.052652 (rank : 37) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y4K4, Q8IYF6 | Gene names | MAP4K5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 5 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 5) (MEK kinase kinase 5) (MEKKK 5) (Kinase homologous to SPS1/STE20) (KHS). | |||||
|
M4K2_HUMAN
|
||||||
| NC score | 0.052455 (rank : 38) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q12851 | Gene names | MAP4K2, GCK, RAB8IP | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 2 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 2) (MEK kinase kinase 2) (MEKKK 2) (Germinal center kinase) (GC kinase) (Rab8-interacting protein) (B lymphocyte serine/threonine-protein kinase). | |||||
|
KPCT_HUMAN
|
||||||
| NC score | 0.052363 (rank : 39) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q04759, Q3MJF1, Q64FY5, Q9H508, Q9H549 | Gene names | PRKCQ, PRKCT | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||
|
TF2AY_MOUSE
|
||||||
| NC score | 0.052056 (rank : 40) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4I4, Q99PM2, Q9DAF4 | Gene names | Gtf2a1lf, Alf | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
KPCD_HUMAN
|
||||||
| NC score | 0.051620 (rank : 41) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q05655, Q15144 | Gene names | PRKCD | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C delta type (EC 2.7.11.13) (nPKC-delta). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.051177 (rank : 42) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
KPCD_MOUSE
|
||||||
| NC score | 0.050967 (rank : 43) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P28867, Q91V85, Q9Z333 | Gene names | Prkcd, Pkcd | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C delta type (EC 2.7.11.13) (nPKC-delta). | |||||
|
CHK1_MOUSE
|
||||||
| NC score | 0.050735 (rank : 44) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O35280, O54925, Q8CI40, Q9D0N2 | Gene names | Chek1, Chk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Chk1 (EC 2.7.11.1). | |||||
|
KPCT_MOUSE
|
||||||
| NC score | 0.050693 (rank : 45) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q02111 | Gene names | Prkcq, Pkcq | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.050185 (rank : 46) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
KKCC2_MOUSE
|
||||||
| NC score | 0.050087 (rank : 47) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8C078, Q80TS0, Q8BXM8, Q8C0G3, Q8CH42, Q8QZT7 | Gene names | Camkk2, Kiaa0787 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 2 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase beta) (CaM-kinase kinase beta) (CaM-KK beta) (CaMKK beta). | |||||
|
KKCC2_HUMAN
|
||||||
| NC score | 0.050050 (rank : 48) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96RR4, O94883, Q8IUG2, Q8IUG3, Q8N3I4, Q8WY03, Q8WY04, Q8WY05, Q8WY06, Q96RP1, Q96RP2, Q96RR3, Q9BWE9, Q9UER3, Q9UES2, Q9Y5N2 | Gene names | CAMKK2, CAMKKB, KIAA0787 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 2 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase beta) (CaM-kinase kinase beta) (CaM-KK beta) (CaMKK beta). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | 0.049977 (rank : 49) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MAST3_HUMAN
|
||||||
| NC score | 0.049519 (rank : 50) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
ST32B_HUMAN
|
||||||
| NC score | 0.049017 (rank : 51) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NY57, Q6UXH3, Q8IY14 | Gene names | STK32B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 32B (EC 2.7.11.1) (YANK2). | |||||
|
ASPX_MOUSE
|
||||||
| NC score | 0.048788 (rank : 52) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50289, Q9DAM6 | Gene names | Acrv1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1) (MSA- 63). | |||||
|
ST32B_MOUSE
|
||||||
| NC score | 0.048706 (rank : 53) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JJX8, Q7TMD3, Q8C4E0 | Gene names | Stk32b, Stk32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 32B (EC 2.7.11.1). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.048409 (rank : 54) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
KS6B2_MOUSE
|
||||||
| NC score | 0.048311 (rank : 55) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z1M4 | Gene names | Rps6kb2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K-beta 2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (S6K2). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.047991 (rank : 56) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
ST32A_HUMAN
|
||||||
| NC score | 0.047497 (rank : 57) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 834 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WU08 | Gene names | STK32A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 32A (EC 2.7.11.1) (YANK1). | |||||
|
TSSK4_MOUSE
|
||||||
| NC score | 0.047263 (rank : 58) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D411 | Gene names | Tssk4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine/threonine-protein kinase 4 (EC 2.7.11.1) (TSSK- 4) (Testis-specific kinase 4) (TSK-4). | |||||
|
EXOC2_MOUSE
|
||||||
| NC score | 0.046989 (rank : 59) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D4H1 | Gene names | Exoc2, Sec5, Sec5l1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
HUNK_MOUSE
|
||||||
| NC score | 0.046898 (rank : 60) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O88866 | Gene names | Hunk, Makv | |||
|
Domain Architecture |

|
|||||
| Description | Hormonally up-regulated neu tumor-associated kinase (EC 2.7.11.1) (Serine/threonine-protein kinase MAK-V). | |||||
|
HUNK_HUMAN
|
||||||
| NC score | 0.046398 (rank : 61) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P57058 | Gene names | HUNK, MAKV | |||
|
Domain Architecture |

|
|||||
| Description | Hormonally up-regulated neu tumor-associated kinase (EC 2.7.11.1) (Serine/threonine-protein kinase MAK-V) (B19). | |||||
|
ST32A_MOUSE
|
||||||
| NC score | 0.046388 (rank : 62) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BGW6, Q7TPQ4 | Gene names | Stk32a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 32A (EC 2.7.11.1). | |||||
|
TTK_MOUSE
|
||||||
| NC score | 0.046166 (rank : 63) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P35761 | Gene names | Ttk, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (ESK) (PYT). | |||||
|
CHK1_HUMAN
|
||||||
| NC score | 0.045545 (rank : 64) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14757 | Gene names | CHEK1, CHK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Chk1 (EC 2.7.11.1). | |||||
|
TTK_HUMAN
|
||||||
| NC score | 0.045516 (rank : 65) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P33981 | Gene names | TTK, MPS1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (Phosphotyrosine picked threonine-protein kinase) (PYT). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.045507 (rank : 66) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
PAK7_MOUSE
|
||||||
| NC score | 0.045445 (rank : 67) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8C015, Q3TQJ7, Q6RWS7 | Gene names | Pak7, Pak5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (p21-activated kinase 5). | |||||
|
STK39_MOUSE
|
||||||
| NC score | 0.045250 (rank : 68) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |

|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
MP2K7_HUMAN
|
||||||
| NC score | 0.045063 (rank : 69) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14733, O14648, O14816, O60452, O60453 | Gene names | MAP2K7, JNKK2, MKK7, PRKMK7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity mitogen-activated protein kinase kinase 7 (EC 2.7.12.2) (MAP kinase kinase 7) (MAPKK 7) (MAPK/ERK kinase 7) (JNK-activating kinase 2) (c-Jun N-terminal kinase kinase 2) (JNK kinase 2) (JNKK 2). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.044636 (rank : 70) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.044256 (rank : 71) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
TAOK3_HUMAN
|
||||||
| NC score | 0.043705 (rank : 72) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1494 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9H2K8, Q658N1, Q8IUM4, Q9HC79, Q9NZM9, Q9UHG7 | Gene names | TAOK3, DPK, JIK, KDS, MAP3K18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3) (Jun kinase-inhibitory kinase) (JNK/SAPK- inhibitory kinase) (Dendritic cell-derived protein kinase) (Cutaneous T-cell lymphoma tumor antigen HD-CL-09) (CTCL tumor antigen HD-CL-09) (Kinase from chicken homolog A) (hKFC-A). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.042689 (rank : 73) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
EXOC2_HUMAN
|
||||||
| NC score | 0.040932 (rank : 74) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KP1, Q5JPC8, Q96AN6, Q9NUZ8, Q9UJM7 | Gene names | EXOC2, SEC5, SEC5L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
SCG1_HUMAN
|
||||||
| NC score | 0.039672 (rank : 75) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.039348 (rank : 76) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.039307 (rank : 77) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
LAS1L_HUMAN
|
||||||
| NC score | 0.038834 (rank : 78) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4W2, Q5JXQ0, Q8TEN5, Q9H9V5 | Gene names | LAS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LAS1-like protein. | |||||
|
CDK7_MOUSE
|
||||||
| NC score | 0.037834 (rank : 79) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q03147, Q99KK3 | Gene names | Cdk7, Cdkn7, Crk4, Mo15, Mpk-7 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division protein kinase 7 (EC 2.7.11.22) (EC 2.7.11.23) (CDK- activating kinase) (CAK) (TFIIH basal transcription factor complex kinase subunit) (39 kDa protein kinase) (P39 Mo15) (Protein-tyrosine kinase MPK-7) (CR4 protein kinase) (CRK4). | |||||
|
CDK7_HUMAN
|
||||||
| NC score | 0.037670 (rank : 80) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P50613, Q9BS60, Q9UE19 | Gene names | CDK7, MO15 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division protein kinase 7 (EC 2.7.11.22) (EC 2.7.11.23) (CDK- activating kinase) (CAK) (TFIIH basal transcription factor complex kinase subunit) (39 kDa protein kinase) (P39 Mo15) (STK1) (CAK1). | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | 0.037359 (rank : 81) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.037116 (rank : 82) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.035963 (rank : 83) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.035390 (rank : 84) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
TNNT3_MOUSE
|
||||||
| NC score | 0.033604 (rank : 85) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.033393 (rank : 86) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.032311 (rank : 87) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
TAI12_HUMAN
|
||||||
| NC score | 0.031695 (rank : 88) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
SSRP1_HUMAN
|
||||||
| NC score | 0.031264 (rank : 89) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
TAI12_MOUSE
|
||||||
| NC score | 0.029108 (rank : 90) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGQ2, Q3UIH1 | Gene names | Taip12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.028661 (rank : 91) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
GP158_MOUSE
|
||||||
| NC score | 0.028201 (rank : 92) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.027824 (rank : 93) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CJ078_HUMAN
|
||||||
| NC score | 0.026923 (rank : 94) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86XK3, Q5JT40, Q8WW47 | Gene names | C10orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78. | |||||
|
TNNT3_HUMAN
|
||||||
| NC score | 0.026654 (rank : 95) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P45378, Q12975, Q12976, Q12977, Q12978, Q86TH6 | Gene names | TNNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT) (Beta TnTF). | |||||
|
MAVS_MOUSE
|
||||||
| NC score | 0.025492 (rank : 96) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
CF153_HUMAN
|
||||||
| NC score | 0.025160 (rank : 97) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96EU6, Q9BRF6, Q9P0C8 | Gene names | C6orf153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf153. | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.024580 (rank : 98) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CO5_MOUSE
|
||||||
| NC score | 0.024462 (rank : 99) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06684 | Gene names | C5, Hc | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor (Hemolytic complement) [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
CF153_MOUSE
|
||||||
| NC score | 0.023993 (rank : 100) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UFY0, Q91X71 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf153 homolog. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.023391 (rank : 101) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.023324 (rank : 102) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.022957 (rank : 103) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.020977 (rank : 104) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.019965 (rank : 105) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.019640 (rank : 106) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.019284 (rank : 107) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
VPS54_MOUSE
|
||||||
| NC score | 0.017216 (rank : 108) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SPW0, Q8BPB3, Q8CFZ7, Q8CHL5, Q8R3R4, Q8R3X1 | Gene names | Vps54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 54 (Tumor antigen SLP-8p homolog). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.015918 (rank : 109) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.015870 (rank : 110) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
LNP_HUMAN
|
||||||
| NC score | 0.015566 (rank : 111) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9C0E8, Q2M2V8, Q2YD99, Q658W8, Q8N5V9, Q96MS5 | Gene names | LNP, KIAA1715 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark. | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.014919 (rank : 112) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.014866 (rank : 113) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.014754 (rank : 114) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
HNRPU_HUMAN
|
||||||
| NC score | 0.014361 (rank : 115) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.013764 (rank : 116) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.013545 (rank : 117) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
FBXW7_HUMAN
|
||||||
| NC score | 0.012333 (rank : 118) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.012209 (rank : 119) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.012067 (rank : 120) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.011669 (rank : 121) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.009124 (rank : 122) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.008764 (rank : 123) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
RBM28_HUMAN
|
||||||
| NC score | 0.008477 (rank : 124) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NW13, Q96CV3 | Gene names | RBM28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
PLXB3_MOUSE
|
||||||
| NC score | 0.005109 (rank : 125) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
HXC10_MOUSE
|
||||||
| NC score | 0.004453 (rank : 126) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P31257, P31312, Q7TMT7 | Gene names | Hoxc10, Hox-3.6, Hoxc-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein Hox-C10 (Hox-3.6). | |||||
|
PTPRG_HUMAN
|
||||||
| NC score | 0.003355 (rank : 127) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
CCKAR_MOUSE
|
||||||
| NC score | 0.001141 (rank : 128) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08786, Q8VCC7, Q9DBV6 | Gene names | Cckar | |||
|
Domain Architecture |

|
|||||
| Description | Cholecystokinin type A receptor (CCK-A receptor) (CCK-AR) (Cholecystokinin-1 receptor) (CCK1-R). | |||||