Please be patient as the page loads
|
LAEVR_HUMAN
|
||||||
| SwissProt Accessions | Q6Q4G3, Q32MR1, Q4G0I9, Q4G0V2, Q86XA3, Q8NBZ2 | Gene names | LVRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-) (CHL2 antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LAEVR_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6Q4G3, Q32MR1, Q4G0I9, Q4G0V2, Q86XA3, Q8NBZ2 | Gene names | LVRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-) (CHL2 antigen). | |||||
|
LAEVR_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989426 (rank : 2) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q2KHK3, Q9D633 | Gene names | Lvrn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-). | |||||
|
AMPN_MOUSE
|
||||||
| θ value | 7.76955e-161 (rank : 3) | NC score | 0.976834 (rank : 3) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97449, Q91YH8, Q99K63 | Gene names | Anpep, Lap-1, Lap1 | |||
|
Domain Architecture |
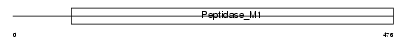
|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (mAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (Membrane protein p161) (CD13 antigen). | |||||
|
AMPN_HUMAN
|
||||||
| θ value | 2.95243e-160 (rank : 4) | NC score | 0.976556 (rank : 4) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15144, Q16728, Q8IUK3, Q8IVH3, Q9UCE0 | Gene names | ANPEP, APN, CD13, PEPN | |||
|
Domain Architecture |
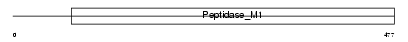
|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (hAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (gp150) (Myeloid plasma membrane glycoprotein CD13) (CD13 antigen). | |||||
|
TRHDE_MOUSE
|
||||||
| θ value | 3.1838e-130 (rank : 5) | NC score | 0.964362 (rank : 6) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K093 | Gene names | Trhde | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
TRHDE_HUMAN
|
||||||
| θ value | 3.5201e-129 (rank : 6) | NC score | 0.965564 (rank : 5) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKU6, Q6UWJ4 | Gene names | TRHDE | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
AMPE_HUMAN
|
||||||
| θ value | 2.14053e-118 (rank : 7) | NC score | 0.963031 (rank : 7) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07075 | Gene names | ENPEP | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (Differentiation antigen gp160) (CD249 antigen). | |||||
|
ARTS1_MOUSE
|
||||||
| θ value | 2.9938e-112 (rank : 8) | NC score | 0.961569 (rank : 10) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EQH2, Q9ET63 | Gene names | Arts1, Appils | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (VEGF-induced aminopeptidase). | |||||
|
AMPE_MOUSE
|
||||||
| θ value | 5.10665e-112 (rank : 9) | NC score | 0.961870 (rank : 8) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16406 | Gene names | Enpep | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (BP-1/6C3 antigen). | |||||
|
ARTS1_HUMAN
|
||||||
| θ value | 3.65967e-110 (rank : 10) | NC score | 0.960768 (rank : 11) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZ08, O60278, Q6UWY6, Q8NEL4, Q8TAD0, Q9UHF8, Q9UKY2 | Gene names | ARTS1, APPILS, KIAA0525 | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator). | |||||
|
LCAP_HUMAN
|
||||||
| θ value | 2.62879e-100 (rank : 11) | NC score | 0.961729 (rank : 9) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UIQ6, O00769, Q15145, Q9UIQ7 | Gene names | LNPEP, OTASE | |||
|
Domain Architecture |
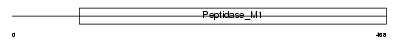
|
|||||
| Description | Leucyl-cystinyl aminopeptidase (EC 3.4.11.3) (Cystinyl aminopeptidase) (Oxytocinase) (OTase) (Insulin-regulated membrane aminopeptidase) (Insulin-responsive aminopeptidase) (IRAP) (Placental leucine aminopeptidase) (P-LAP). | |||||
|
PSA_HUMAN
|
||||||
| θ value | 3.0217e-80 (rank : 12) | NC score | 0.946002 (rank : 12) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55786, Q6P145, Q9NP16, Q9UEM2 | Gene names | NPEPPS, PSA | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
PSA_MOUSE
|
||||||
| θ value | 5.15425e-80 (rank : 13) | NC score | 0.945991 (rank : 13) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q11011, Q91VJ8 | Gene names | Npepps, Psa | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
LKHA4_MOUSE
|
||||||
| θ value | 2.20094e-22 (rank : 14) | NC score | 0.716177 (rank : 15) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P24527, Q8VDR8 | Gene names | Lta4h | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
LKHA4_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 15) | NC score | 0.723501 (rank : 14) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09960 | Gene names | LTA4H, LTA4 | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
AMPB_MOUSE
|
||||||
| θ value | 1.02238e-19 (rank : 16) | NC score | 0.681396 (rank : 16) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VCT3 | Gene names | Rnpep | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase) (Cytosol aminopeptidase IV). | |||||
|
AMPB_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 17) | NC score | 0.668807 (rank : 17) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H4A4, Q9BVM9, Q9H1D4, Q9NPT7 | Gene names | RNPEP, APB | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase). | |||||
|
RNPL1_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 18) | NC score | 0.647211 (rank : 18) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAU8, Q96AC9, Q9H033, Q9NVD0 | Gene names | RNPEPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl aminopeptidase-like 1 (EC 3.4.11.-) (RNPEP-like protein). | |||||
|
AMPO_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 19) | NC score | 0.461076 (rank : 19) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N6M6, Q5T9B1, Q5T9B3, Q5T9B4, Q8WUL6, Q96M23, Q96SS1 | Gene names | ONPEP, C9orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
AMPO_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 20) | NC score | 0.446060 (rank : 20) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BXQ6, Q6P0W6, Q6P394, Q8BHX5 | Gene names | Onpep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.013255 (rank : 35) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.031454 (rank : 24) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
M4K1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | -0.000135 (rank : 73) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92918 | Gene names | MAP4K1, HPK1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 1 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 1) (MEK kinase kinase 1) (MEKKK 1) (Hematopoietic progenitor kinase). | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.019594 (rank : 28) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.008377 (rank : 46) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.008073 (rank : 49) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
CU030_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.037672 (rank : 23) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UFM2 | Gene names | C21orf30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C21orf30. | |||||
|
IGS4B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.011131 (rank : 40) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99N28, Q8BSQ8, Q8K1H8 | Gene names | Igsf4b, Necl1, Syncam3, Tsll1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin superfamily member 4B precursor (Nectin-like protein 1) (Necl-1) (TSLC1-like protein 1) (Synaptic cell adhesion molecule 3). | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.018828 (rank : 29) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.026992 (rank : 25) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.012165 (rank : 38) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.018700 (rank : 30) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.020108 (rank : 27) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.021633 (rank : 26) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.015123 (rank : 33) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.009246 (rank : 44) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.009736 (rank : 42) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
AB1IP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.011576 (rank : 39) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.016537 (rank : 31) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.008298 (rank : 48) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
HXA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.002615 (rank : 69) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.012729 (rank : 36) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.006953 (rank : 57) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
CENG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.003986 (rank : 64) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
DYR1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | -0.001121 (rank : 75) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z188 | Gene names | Dyrk1b | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1B (EC 2.7.12.1). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.006521 (rank : 60) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.007951 (rank : 50) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.006976 (rank : 56) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ADA12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.001814 (rank : 70) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43184, O60470 | Gene names | ADAM12, MLTN | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
CASL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.008735 (rank : 45) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |
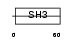
|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.012600 (rank : 37) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.006378 (rank : 61) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.008337 (rank : 47) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
MTLR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | -0.000926 (rank : 74) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43193 | Gene names | MLNR, GPR38, MTLR, MTLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Motilin receptor (G-protein coupled receptor 38). | |||||
|
SF3A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.014823 (rank : 34) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
SF3A1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.015146 (rank : 32) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
CNTP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.003601 (rank : 66) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
COHA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.004713 (rank : 63) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
ES8L3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.006737 (rank : 59) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TE67, Q5T8Q6, Q5T8Q7, Q5T8Q8, Q96E47, Q9H719 | Gene names | EPS8L3, EPS8R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.007210 (rank : 54) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
GCS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.009297 (rank : 43) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UM7, Q9Z2W5 | Gene names | Gcs1 | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide glucosidase (EC 3.2.1.106) (Glycoprotein- processing glucosidase I) (Glucosidase 1). | |||||
|
IGS4B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.007460 (rank : 52) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N126, Q9NVJ5, Q9UJP1 | Gene names | IGSF4B, NECL1, SYNCAM3, TSLL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin superfamily member 4B precursor (Nectin-like protein 1) (TSLC1-like protein 1) (Synaptic cell adhesion molecule 3) (Brain immunoglobulin receptor). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.002653 (rank : 68) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.003959 (rank : 65) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SF3B3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.010700 (rank : 41) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15393, Q9UJ29 | Gene names | SF3B3, KIAA0017, SAP130 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor 3B subunit 3 (Spliceosome-associated protein 130) (SAP 130) (SF3b130) (Pre-mRNA-splicing factor SF3b 130 kDa subunit) (STAF130). | |||||
|
SYN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.007824 (rank : 51) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.006366 (rank : 62) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ABI3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.007381 (rank : 53) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2A4, Q9H0P6 | Gene names | ABI3, NESH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.001375 (rank : 72) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
FREA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.001547 (rank : 71) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.006828 (rank : 58) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.007057 (rank : 55) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ZYX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.003446 (rank : 67) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62523, P70461 | Gene names | Zyx | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin. | |||||
|
TAF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.161782 (rank : 21) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P1X5, O43487, O43604, O60668, Q86WW7, Q8IWK4 | Gene names | TAF2, CIF150, TAF2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa) (TAFII-150) (TAFII150) (150 kDa cofactor of initiator function). | |||||
|
TAF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.153786 (rank : 22) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C176, Q8BKQ7 | Gene names | Taf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa). | |||||
|
LAEVR_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6Q4G3, Q32MR1, Q4G0I9, Q4G0V2, Q86XA3, Q8NBZ2 | Gene names | LVRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-) (CHL2 antigen). | |||||
|
LAEVR_MOUSE
|
||||||
| NC score | 0.989426 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q2KHK3, Q9D633 | Gene names | Lvrn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-). | |||||
|
AMPN_MOUSE
|
||||||
| NC score | 0.976834 (rank : 3) | θ value | 7.76955e-161 (rank : 3) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97449, Q91YH8, Q99K63 | Gene names | Anpep, Lap-1, Lap1 | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (mAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (Membrane protein p161) (CD13 antigen). | |||||
|
AMPN_HUMAN
|
||||||
| NC score | 0.976556 (rank : 4) | θ value | 2.95243e-160 (rank : 4) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15144, Q16728, Q8IUK3, Q8IVH3, Q9UCE0 | Gene names | ANPEP, APN, CD13, PEPN | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (hAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (gp150) (Myeloid plasma membrane glycoprotein CD13) (CD13 antigen). | |||||
|
TRHDE_HUMAN
|
||||||
| NC score | 0.965564 (rank : 5) | θ value | 3.5201e-129 (rank : 6) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKU6, Q6UWJ4 | Gene names | TRHDE | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
TRHDE_MOUSE
|
||||||
| NC score | 0.964362 (rank : 6) | θ value | 3.1838e-130 (rank : 5) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K093 | Gene names | Trhde | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
AMPE_HUMAN
|
||||||
| NC score | 0.963031 (rank : 7) | θ value | 2.14053e-118 (rank : 7) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07075 | Gene names | ENPEP | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (Differentiation antigen gp160) (CD249 antigen). | |||||
|
AMPE_MOUSE
|
||||||
| NC score | 0.961870 (rank : 8) | θ value | 5.10665e-112 (rank : 9) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16406 | Gene names | Enpep | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (BP-1/6C3 antigen). | |||||
|
LCAP_HUMAN
|
||||||
| NC score | 0.961729 (rank : 9) | θ value | 2.62879e-100 (rank : 11) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UIQ6, O00769, Q15145, Q9UIQ7 | Gene names | LNPEP, OTASE | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-cystinyl aminopeptidase (EC 3.4.11.3) (Cystinyl aminopeptidase) (Oxytocinase) (OTase) (Insulin-regulated membrane aminopeptidase) (Insulin-responsive aminopeptidase) (IRAP) (Placental leucine aminopeptidase) (P-LAP). | |||||
|
ARTS1_MOUSE
|
||||||
| NC score | 0.961569 (rank : 10) | θ value | 2.9938e-112 (rank : 8) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EQH2, Q9ET63 | Gene names | Arts1, Appils | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (VEGF-induced aminopeptidase). | |||||
|
ARTS1_HUMAN
|
||||||
| NC score | 0.960768 (rank : 11) | θ value | 3.65967e-110 (rank : 10) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZ08, O60278, Q6UWY6, Q8NEL4, Q8TAD0, Q9UHF8, Q9UKY2 | Gene names | ARTS1, APPILS, KIAA0525 | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator). | |||||
|
PSA_HUMAN
|
||||||
| NC score | 0.946002 (rank : 12) | θ value | 3.0217e-80 (rank : 12) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55786, Q6P145, Q9NP16, Q9UEM2 | Gene names | NPEPPS, PSA | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
PSA_MOUSE
|
||||||
| NC score | 0.945991 (rank : 13) | θ value | 5.15425e-80 (rank : 13) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q11011, Q91VJ8 | Gene names | Npepps, Psa | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
LKHA4_HUMAN
|
||||||
| NC score | 0.723501 (rank : 14) | θ value | 4.15078e-21 (rank : 15) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09960 | Gene names | LTA4H, LTA4 | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
LKHA4_MOUSE
|
||||||
| NC score | 0.716177 (rank : 15) | θ value | 2.20094e-22 (rank : 14) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P24527, Q8VDR8 | Gene names | Lta4h | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
AMPB_MOUSE
|
||||||
| NC score | 0.681396 (rank : 16) | θ value | 1.02238e-19 (rank : 16) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VCT3 | Gene names | Rnpep | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase) (Cytosol aminopeptidase IV). | |||||
|
AMPB_HUMAN
|
||||||
| NC score | 0.668807 (rank : 17) | θ value | 1.52774e-15 (rank : 17) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H4A4, Q9BVM9, Q9H1D4, Q9NPT7 | Gene names | RNPEP, APB | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase). | |||||
|
RNPL1_HUMAN
|
||||||
| NC score | 0.647211 (rank : 18) | θ value | 6.00763e-12 (rank : 18) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAU8, Q96AC9, Q9H033, Q9NVD0 | Gene names | RNPEPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl aminopeptidase-like 1 (EC 3.4.11.-) (RNPEP-like protein). | |||||
|
AMPO_HUMAN
|
||||||
| NC score | 0.461076 (rank : 19) | θ value | 2.44474e-05 (rank : 19) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N6M6, Q5T9B1, Q5T9B3, Q5T9B4, Q8WUL6, Q96M23, Q96SS1 | Gene names | ONPEP, C9orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
AMPO_MOUSE
|
||||||
| NC score | 0.446060 (rank : 20) | θ value | 4.1701e-05 (rank : 20) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BXQ6, Q6P0W6, Q6P394, Q8BHX5 | Gene names | Onpep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
TAF2_HUMAN
|
||||||
| NC score | 0.161782 (rank : 21) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P1X5, O43487, O43604, O60668, Q86WW7, Q8IWK4 | Gene names | TAF2, CIF150, TAF2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa) (TAFII-150) (TAFII150) (150 kDa cofactor of initiator function). | |||||
|
TAF2_MOUSE
|
||||||
| NC score | 0.153786 (rank : 22) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C176, Q8BKQ7 | Gene names | Taf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa). | |||||
|
CU030_HUMAN
|
||||||
| NC score | 0.037672 (rank : 23) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UFM2 | Gene names | C21orf30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C21orf30. | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.031454 (rank : 24) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.026992 (rank : 25) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.021633 (rank : 26) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.020108 (rank : 27) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.019594 (rank : 28) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.018828 (rank : 29) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.018700 (rank : 30) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.016537 (rank : 31) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SF3A1_MOUSE
|
||||||
| NC score | 0.015146 (rank : 32) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.015123 (rank : 33) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SF3A1_HUMAN
|
||||||
| NC score | 0.014823 (rank : 34) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.013255 (rank : 35) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.012729 (rank : 36) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.012600 (rank : 37) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.012165 (rank : 38) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
AB1IP_HUMAN
|
||||||
| NC score | 0.011576 (rank : 39) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
IGS4B_MOUSE
|
||||||
| NC score | 0.011131 (rank : 40) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99N28, Q8BSQ8, Q8K1H8 | Gene names | Igsf4b, Necl1, Syncam3, Tsll1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin superfamily member 4B precursor (Nectin-like protein 1) (Necl-1) (TSLC1-like protein 1) (Synaptic cell adhesion molecule 3). | |||||
|
SF3B3_HUMAN
|
||||||
| NC score | 0.010700 (rank : 41) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15393, Q9UJ29 | Gene names | SF3B3, KIAA0017, SAP130 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 3 (Spliceosome-associated protein 130) (SAP 130) (SF3b130) (Pre-mRNA-splicing factor SF3b 130 kDa subunit) (STAF130). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.009736 (rank : 42) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
GCS1_MOUSE
|
||||||
| NC score | 0.009297 (rank : 43) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UM7, Q9Z2W5 | Gene names | Gcs1 | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide glucosidase (EC 3.2.1.106) (Glycoprotein- processing glucosidase I) (Glucosidase 1). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.009246 (rank : 44) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
CASL_MOUSE
|
||||||
| NC score | 0.008735 (rank : 45) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.008377 (rank : 46) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.008337 (rank : 47) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.008298 (rank : 48) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.008073 (rank : 49) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.007951 (rank : 50) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SYN2_MOUSE
|
||||||
| NC score | 0.007824 (rank : 51) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
IGS4B_HUMAN
|
||||||
| NC score | 0.007460 (rank : 52) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N126, Q9NVJ5, Q9UJP1 | Gene names | IGSF4B, NECL1, SYNCAM3, TSLL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin superfamily member 4B precursor (Nectin-like protein 1) (TSLC1-like protein 1) (Synaptic cell adhesion molecule 3) (Brain immunoglobulin receptor). | |||||
|
ABI3_HUMAN
|
||||||
| NC score | 0.007381 (rank : 53) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2A4, Q9H0P6 | Gene names | ABI3, NESH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.007210 (rank : 54) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.007057 (rank : 55) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.006976 (rank : 56) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.006953 (rank : 57) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.006828 (rank : 58) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
ES8L3_HUMAN
|
||||||
| NC score | 0.006737 (rank : 59) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TE67, Q5T8Q6, Q5T8Q7, Q5T8Q8, Q96E47, Q9H719 | Gene names | EPS8L3, EPS8R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.006521 (rank : 60) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.006378 (rank : 61) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.006366 (rank : 62) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.004713 (rank : 63) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
CENG1_HUMAN
|
||||||
| NC score | 0.003986 (rank : 64) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.003959 (rank : 65) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
CNTP1_HUMAN
|
||||||
| NC score | 0.003601 (rank : 66) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
ZYX_MOUSE
|
||||||
| NC score | 0.003446 (rank : 67) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62523, P70461 | Gene names | Zyx | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin. | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.002653 (rank : 68) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
HXA3_MOUSE
|
||||||
| NC score | 0.002615 (rank : 69) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
ADA12_HUMAN
|
||||||
| NC score | 0.001814 (rank : 70) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43184, O60470 | Gene names | ADAM12, MLTN | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
FREA_HUMAN
|
||||||
| NC score | 0.001547 (rank : 71) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.001375 (rank : 72) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
M4K1_HUMAN
|
||||||
| NC score | -0.000135 (rank : 73) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92918 | Gene names | MAP4K1, HPK1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 1 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 1) (MEK kinase kinase 1) (MEKKK 1) (Hematopoietic progenitor kinase). | |||||
|
MTLR_HUMAN
|
||||||
| NC score | -0.000926 (rank : 74) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43193 | Gene names | MLNR, GPR38, MTLR, MTLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Motilin receptor (G-protein coupled receptor 38). | |||||
|
DYR1B_MOUSE
|
||||||
| NC score | -0.001121 (rank : 75) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z188 | Gene names | Dyrk1b | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1B (EC 2.7.12.1). | |||||