Please be patient as the page loads
|
HDAC6_HUMAN
|
||||||
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HDAC6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
HDAC6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.979548 (rank : 2) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
HDA10_HUMAN
|
||||||
| θ value | 3.53647e-113 (rank : 3) | NC score | 0.921519 (rank : 4) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q969S8, Q6STF9, Q96P77, Q96P78, Q9H028, Q9UGX1, Q9UGX2 | Gene names | HDAC10 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 10 (HD10). | |||||
|
HDA10_MOUSE
|
||||||
| θ value | 7.37397e-111 (rank : 4) | NC score | 0.921801 (rank : 3) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P3E7 | Gene names | Hdac10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone deacetylase 10 (HD10). | |||||
|
HDAC4_HUMAN
|
||||||
| θ value | 9.39325e-82 (rank : 5) | NC score | 0.817159 (rank : 9) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
HDAC7_MOUSE
|
||||||
| θ value | 3.23591e-74 (rank : 6) | NC score | 0.873189 (rank : 5) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
HDAC5_MOUSE
|
||||||
| θ value | 2.09745e-73 (rank : 7) | NC score | 0.821975 (rank : 7) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
HDAC7_HUMAN
|
||||||
| θ value | 2.09745e-73 (rank : 8) | NC score | 0.871394 (rank : 6) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WUI4, Q7Z5I1, Q96K01, Q9BR73, Q9H7L0, Q9NW41, Q9NWA9, Q9NYK9, Q9UFU7 | Gene names | HDAC7A, HDAC7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
HDAC5_HUMAN
|
||||||
| θ value | 3.57772e-73 (rank : 9) | NC score | 0.809548 (rank : 10) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
HDAC9_HUMAN
|
||||||
| θ value | 8.81223e-72 (rank : 10) | NC score | 0.818884 (rank : 8) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UKV0, O94845, O95028 | Gene names | HDAC9, HDAC7, HDAC7B, KIAA0744 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (HD7). | |||||
|
HDAC8_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 11) | NC score | 0.715923 (rank : 11) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BY41, Q9NP76, Q9NYH4 | Gene names | HDAC8, HDACL1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 8 (HD8). | |||||
|
HDAC8_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 12) | NC score | 0.713502 (rank : 12) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VH37, Q3V270, Q9D0K6 | Gene names | Hdac8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 8 (HD8). | |||||
|
HDAC2_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 13) | NC score | 0.683459 (rank : 15) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92769, Q5SRI8, Q5SZ86, Q8NEH4 | Gene names | HDAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 2 (HD2). | |||||
|
HDAC1_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 14) | NC score | 0.684692 (rank : 13) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13547, Q92534 | Gene names | HDAC1, RPD3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 1 (HD1). | |||||
|
HDAC1_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 15) | NC score | 0.684510 (rank : 14) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09106, P97476 | Gene names | Hdac1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 1 (HD1). | |||||
|
HDAC2_MOUSE
|
||||||
| θ value | 1.86321e-21 (rank : 16) | NC score | 0.682542 (rank : 16) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70288 | Gene names | Hdac2, Yy1bp | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 2 (HD2) (YY1 transcription factor-binding protein). | |||||
|
HDAC3_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 17) | NC score | 0.651393 (rank : 18) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88895, O88896 | Gene names | Hdac3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 3 (HD3). | |||||
|
HDAC3_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 18) | NC score | 0.650385 (rank : 19) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15379, O43268, Q9UEI5, Q9UEV0 | Gene names | HDAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 3 (HD3) (RPD3-2) (SMAP45). | |||||
|
HDA11_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 19) | NC score | 0.666529 (rank : 17) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96DB2, Q9H6I7, Q9H6X3, Q9NTC9 | Gene names | HDAC11 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 11 (HD11). | |||||
|
HDA11_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 20) | NC score | 0.648329 (rank : 20) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91WA3 | Gene names | Hdac11 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 11 (HD11). | |||||
|
UBP49_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 21) | NC score | 0.211975 (rank : 22) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 22) | NC score | 0.211528 (rank : 23) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 23) | NC score | 0.207116 (rank : 24) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 24) | NC score | 0.108704 (rank : 33) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
UBP3_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 25) | NC score | 0.163213 (rank : 28) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP3_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 26) | NC score | 0.167351 (rank : 26) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
BRAP_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 27) | NC score | 0.166363 (rank : 27) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
BRAP_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 28) | NC score | 0.189013 (rank : 25) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
UBP20_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 29) | NC score | 0.145716 (rank : 29) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 30) | NC score | 0.063818 (rank : 63) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SELPL_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 31) | NC score | 0.102047 (rank : 35) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
UBP13_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 32) | NC score | 0.119180 (rank : 32) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 33) | NC score | 0.131038 (rank : 31) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
OVGP1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 34) | NC score | 0.032334 (rank : 107) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62010 | Gene names | Ovgp1, Chit5, Ogp | |||
|
Domain Architecture |

|
|||||
| Description | Oviduct-specific glycoprotein precursor (Oviductal glycoprotein) (Oviductin) (Estrogen-dependent oviduct protein). | |||||
|
NUP62_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 35) | NC score | 0.071179 (rank : 51) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 36) | NC score | 0.131199 (rank : 30) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 37) | NC score | 0.040490 (rank : 100) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 38) | NC score | 0.052377 (rank : 80) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 39) | NC score | 0.010529 (rank : 154) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 40) | NC score | 0.044188 (rank : 91) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PME17_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 41) | NC score | 0.056427 (rank : 78) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
PME17_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 42) | NC score | 0.067651 (rank : 54) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40967, Q12763, Q14448, Q14817, Q16565 | Gene names | SILV, D12S53E, PMEL17 | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Melanocyte lineage-specific antigen GP100) (Melanoma-associated ME20 antigen) (ME20M/ME20S) (ME20- M/ME20-S) (95 kDa melanocyte-specific secreted glycoprotein). | |||||
|
K0240_HUMAN
|
||||||
| θ value | 0.125558 (rank : 43) | NC score | 0.037355 (rank : 104) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 44) | NC score | 0.066990 (rank : 57) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 45) | NC score | 0.099852 (rank : 37) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 46) | NC score | 0.100084 (rank : 36) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 47) | NC score | 0.042056 (rank : 95) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 0.163984 (rank : 48) | NC score | 0.059538 (rank : 67) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
CAMKV_MOUSE
|
||||||
| θ value | 0.21417 (rank : 49) | NC score | 0.004165 (rank : 175) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UHL1, Q8VD20 | Gene names | Camkv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
BASP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 50) | NC score | 0.043207 (rank : 92) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.056793 (rank : 77) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
NFYA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 52) | NC score | 0.042908 (rank : 93) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23511, Q8IXU0 | Gene names | NFYA | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit alpha (Nuclear transcription factor Y subunit A) (NF-YA) (CAAT-box DNA-binding protein subunit A). | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.017560 (rank : 135) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
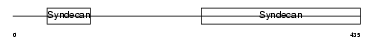
SDC3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 54) | NC score | 0.042182 (rank : 94) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64519, Q3UDD9, Q6ZQA4, Q7TQD4 | Gene names | Sdc3, Kiaa0468 | |||
|
Domain Architecture |
|
|||||
| Description | Syndecan-3 precursor (SYND3). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.016347 (rank : 141) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
CCG8_MOUSE
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.016904 (rank : 140) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHW2 | Gene names | Cacng8 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-8 subunit (Neuronal voltage- gated calcium channel gamma-8 subunit). | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.041286 (rank : 98) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MA2B2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.020101 (rank : 125) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54782, Q69ZV1, Q8BH85, Q9DBK2 | Gene names | Man2b2, Kiaa0935 | |||
|
Domain Architecture |

|
|||||
| Description | Epididymis-specific alpha-mannosidase precursor (EC 3.2.1.24) (Mannosidase alpha class 2B member 2). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.038458 (rank : 102) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.015772 (rank : 143) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.041445 (rank : 97) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
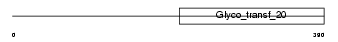
ALG2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.028630 (rank : 111) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBE8, Q7TN30, Q9CWI6 | Gene names | Alg2 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1,3-mannosyltransferase ALG2 (EC 2.4.1.-) (GDP- Man:Man(1)GlcNAc(2)-PP-dolichol mannosyltransferase). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.010986 (rank : 151) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.040632 (rank : 99) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.058008 (rank : 71) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.052016 (rank : 81) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 0.62314 (rank : 67) | NC score | 0.102930 (rank : 34) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 68) | NC score | 0.034685 (rank : 106) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.040359 (rank : 101) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
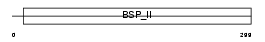
SIAL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.036604 (rank : 105) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61711, Q61363 | Gene names | Ibsp | |||
|
Domain Architecture |
|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
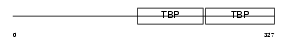
TBP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 71) | NC score | 0.028590 (rank : 112) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20226, Q16845, Q9UC02 | Gene names | TBP, TF2D, TFIID | |||
|
Domain Architecture |
|
|||||
| Description | TATA-box-binding protein (TATA-box factor) (TATA-binding factor) (TATA sequence-binding protein) (Transcription initiation factor TFIID TBP subunit). | |||||
|
NUPL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.027097 (rank : 115) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R332, Q3UJF4, Q8BUA7, Q8BVG7, Q8C0W3 | Gene names | Nupl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin p58/p45 (Nucleoporin-like 1). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.041517 (rank : 96) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.023458 (rank : 119) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.007773 (rank : 164) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.008567 (rank : 160) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
NUP62_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.051122 (rank : 86) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
OTU7A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.017566 (rank : 134) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.001353 (rank : 181) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
T240L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.018923 (rank : 129) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
A4GCT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.027562 (rank : 113) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNA3 | Gene names | A4GNT | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1,4-N-acetylglucosaminyltransferase (EC 2.4.1.-) (Alpha4GnT). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.015203 (rank : 144) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.027388 (rank : 114) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.022150 (rank : 121) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.063922 (rank : 61) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.030328 (rank : 110) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.017399 (rank : 137) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
PTPR2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.005505 (rank : 172) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.021473 (rank : 123) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.030406 (rank : 109) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.020026 (rank : 126) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
DMN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.010003 (rank : 155) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.006451 (rank : 169) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.009780 (rank : 157) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.021364 (rank : 124) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
UBP22_MOUSE
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.093353 (rank : 39) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
ZNF76_HUMAN
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | -0.002891 (rank : 184) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36508, Q9BQB2 | Gene names | ZNF76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 76. | |||||
|
ADNP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.018367 (rank : 130) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H2P0, O94881, Q9UG34 | Gene names | ADNP, KIAA0784 | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.007682 (rank : 165) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CUGB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.010813 (rank : 153) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92879, Q9NP83, Q9NR06 | Gene names | CUGBP1, BRUNOL2, CUGBP, NAB50 | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (50 kDa Nuclear polyadenylated RNA-binding protein) (EDEN-BP). | |||||
|
CUGB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.010825 (rank : 152) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28659, Q9CXE5, Q9EPJ8, Q9JI37 | Gene names | Cugbp1, Brunol2, Cugbp | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (Deadenylation factor EDEN-BP) (Brain protein F41). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.008676 (rank : 159) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
LRBA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.007552 (rank : 166) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
NFRKB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.037438 (rank : 103) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.017647 (rank : 133) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
VASP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.017259 (rank : 139) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
ACD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.017418 (rank : 136) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96AP0, Q562H5, Q9H8F9 | Gene names | ACD, PIP1, PTOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adrenocortical dysplasia protein homolog (POT1 and TIN2-interacting protein). | |||||
|
CF142_MOUSE
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.017368 (rank : 138) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5FW52, Q9D6X9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf142 homolog. | |||||
|
CLIC6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.009683 (rank : 158) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.026391 (rank : 116) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
VASP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.016053 (rank : 142) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
ZN335_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | -0.000874 (rank : 182) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
ASH2L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.011846 (rank : 149) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.021661 (rank : 122) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.025954 (rank : 117) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.013353 (rank : 147) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.031189 (rank : 108) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PORIM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.022657 (rank : 120) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N131, Q8IWS2, Q96QV2 | Gene names | TMEM123, KCT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123) (Pro-oncosis receptor inducing membrane injury) (Keratinocytes-associated transmembrane protein 3) (KCT-3). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.008390 (rank : 162) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
SEM4D_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.003509 (rank : 177) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
UBP22_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.094847 (rank : 38) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.047433 (rank : 90) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.006229 (rank : 170) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
I17RA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.008565 (rank : 161) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96F46, O43844 | Gene names | IL17RA, IL17R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-17 receptor A precursor (IL-17 receptor). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.019784 (rank : 127) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.017932 (rank : 131) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
RFX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.013229 (rank : 148) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
SIAL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.024038 (rank : 118) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |
|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
UBQL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.006741 (rank : 167) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R317, Q80V10, Q8C7T4, Q8C835, Q8K141, Q91VI8, Q9D0Z0, Q9QZM1 | Gene names | Ubqln1, Plic1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-1 (Protein linking IAP with cytoskeleton 1) (PLIC-1). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.004812 (rank : 174) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.019292 (rank : 128) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
AUXI_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.005485 (rank : 173) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.017647 (rank : 132) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BMP5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.001565 (rank : 180) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49003 | Gene names | Bmp5, Bmp-5 | |||
|
Domain Architecture |
|
|||||
| Description | Bone morphogenetic protein 5 precursor (BMP-5). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.014652 (rank : 145) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ICAL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.011836 (rank : 150) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
LGR5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | -0.001443 (rank : 183) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1P4 | Gene names | Lgr5, Fex, Gpr49 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 5 precursor (G-protein coupled receptor 49) (Orphan G-protein coupled receptor FEX). | |||||
|
MAGAB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.005928 (rank : 171) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.002721 (rank : 179) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PHF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.007971 (rank : 163) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.003405 (rank : 178) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
TAOK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.003707 (rank : 176) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
TRBP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.006538 (rank : 168) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.009999 (rank : 156) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
UBN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.013662 (rank : 146) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
CK024_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.051774 (rank : 83) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
HDAC9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.400628 (rank : 21) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99N13, Q9EPT2 | Gene names | Hdac9, Hdac7b, Hdrp, Mitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (Histone deacetylase-related protein) (MEF2-interacting transcription repressor MITR). | |||||
|
SNUT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.050771 (rank : 88) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
SNUT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.051234 (rank : 85) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
SYCP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.051827 (rank : 82) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZU3 | Gene names | SYCP3, SCP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 3 (SCP-3). | |||||
|
UB17L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.064334 (rank : 59) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.055946 (rank : 79) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.058105 (rank : 70) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.057788 (rank : 72) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.068266 (rank : 53) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.057547 (rank : 74) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.057582 (rank : 73) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.056876 (rank : 76) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.057323 (rank : 75) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.072813 (rank : 42) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.072094 (rank : 48) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.072751 (rank : 43) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.072050 (rank : 49) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.072676 (rank : 44) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP30_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.063868 (rank : 62) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.072349 (rank : 45) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.051063 (rank : 87) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.051319 (rank : 84) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.071181 (rank : 50) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP40_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.050072 (rank : 89) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.076405 (rank : 40) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.062979 (rank : 65) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.064282 (rank : 60) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.067403 (rank : 55) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.067403 (rank : 56) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP48_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.062255 (rank : 66) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.063102 (rank : 64) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.059233 (rank : 68) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.058590 (rank : 69) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.072152 (rank : 47) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.075373 (rank : 41) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.070160 (rank : 52) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.072221 (rank : 46) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBPW_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.065652 (rank : 58) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
HDAC6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
HDAC6_MOUSE
|
||||||
| NC score | 0.979548 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
HDA10_MOUSE
|
||||||
| NC score | 0.921801 (rank : 3) | θ value | 7.37397e-111 (rank : 4) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P3E7 | Gene names | Hdac10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone deacetylase 10 (HD10). | |||||
|
HDA10_HUMAN
|
||||||
| NC score | 0.921519 (rank : 4) | θ value | 3.53647e-113 (rank : 3) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q969S8, Q6STF9, Q96P77, Q96P78, Q9H028, Q9UGX1, Q9UGX2 | Gene names | HDAC10 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 10 (HD10). | |||||
|
HDAC7_MOUSE
|
||||||
| NC score | 0.873189 (rank : 5) | θ value | 3.23591e-74 (rank : 6) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
HDAC7_HUMAN
|
||||||
| NC score | 0.871394 (rank : 6) | θ value | 2.09745e-73 (rank : 8) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WUI4, Q7Z5I1, Q96K01, Q9BR73, Q9H7L0, Q9NW41, Q9NWA9, Q9NYK9, Q9UFU7 | Gene names | HDAC7A, HDAC7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
HDAC5_MOUSE
|
||||||
| NC score | 0.821975 (rank : 7) | θ value | 2.09745e-73 (rank : 7) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
HDAC9_HUMAN
|
||||||
| NC score | 0.818884 (rank : 8) | θ value | 8.81223e-72 (rank : 10) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UKV0, O94845, O95028 | Gene names | HDAC9, HDAC7, HDAC7B, KIAA0744 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (HD7). | |||||
|
HDAC4_HUMAN
|
||||||
| NC score | 0.817159 (rank : 9) | θ value | 9.39325e-82 (rank : 5) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
HDAC5_HUMAN
|
||||||
| NC score | 0.809548 (rank : 10) | θ value | 3.57772e-73 (rank : 9) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
HDAC8_HUMAN
|
||||||
| NC score | 0.715923 (rank : 11) | θ value | 2.87452e-22 (rank : 11) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BY41, Q9NP76, Q9NYH4 | Gene names | HDAC8, HDACL1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 8 (HD8). | |||||
|
HDAC8_MOUSE
|
||||||
| NC score | 0.713502 (rank : 12) | θ value | 2.87452e-22 (rank : 12) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VH37, Q3V270, Q9D0K6 | Gene names | Hdac8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 8 (HD8). | |||||
|
HDAC1_HUMAN
|
||||||
| NC score | 0.684692 (rank : 13) | θ value | 1.42661e-21 (rank : 14) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13547, Q92534 | Gene names | HDAC1, RPD3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 1 (HD1). | |||||
|
HDAC1_MOUSE
|
||||||
| NC score | 0.684510 (rank : 14) | θ value | 1.42661e-21 (rank : 15) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09106, P97476 | Gene names | Hdac1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 1 (HD1). | |||||
|
HDAC2_HUMAN
|
||||||
| NC score | 0.683459 (rank : 15) | θ value | 1.09232e-21 (rank : 13) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92769, Q5SRI8, Q5SZ86, Q8NEH4 | Gene names | HDAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 2 (HD2). | |||||
|
HDAC2_MOUSE
|
||||||
| NC score | 0.682542 (rank : 16) | θ value | 1.86321e-21 (rank : 16) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70288 | Gene names | Hdac2, Yy1bp | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 2 (HD2) (YY1 transcription factor-binding protein). | |||||
|
HDA11_HUMAN
|
||||||
| NC score | 0.666529 (rank : 17) | θ value | 3.40345e-15 (rank : 19) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96DB2, Q9H6I7, Q9H6X3, Q9NTC9 | Gene names | HDAC11 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 11 (HD11). | |||||
|
HDAC3_MOUSE
|
||||||
| NC score | 0.651393 (rank : 18) | θ value | 5.25075e-16 (rank : 17) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88895, O88896 | Gene names | Hdac3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 3 (HD3). | |||||
|
HDAC3_HUMAN
|
||||||
| NC score | 0.650385 (rank : 19) | θ value | 6.85773e-16 (rank : 18) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15379, O43268, Q9UEI5, Q9UEV0 | Gene names | HDAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 3 (HD3) (RPD3-2) (SMAP45). | |||||
|
HDA11_MOUSE
|
||||||
| NC score | 0.648329 (rank : 20) | θ value | 1.58096e-12 (rank : 20) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91WA3 | Gene names | Hdac11 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 11 (HD11). | |||||
|
HDAC9_MOUSE
|
||||||
| NC score | 0.400628 (rank : 21) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99N13, Q9EPT2 | Gene names | Hdac9, Hdac7b, Hdrp, Mitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (Histone deacetylase-related protein) (MEF2-interacting transcription repressor MITR). | |||||
|
UBP49_HUMAN
|
||||||
| NC score | 0.211975 (rank : 22) | θ value | 1.74796e-11 (rank : 21) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_MOUSE
|
||||||
| NC score | 0.211528 (rank : 23) | θ value | 2.28291e-11 (rank : 22) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.207116 (rank : 24) | θ value | 3.89403e-11 (rank : 23) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
BRAP_MOUSE
|
||||||
| NC score | 0.189013 (rank : 25) | θ value | 3.77169e-06 (rank : 28) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
UBP3_HUMAN
|
||||||
| NC score | 0.167351 (rank : 26) | θ value | 1.99992e-07 (rank : 26) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
BRAP_HUMAN
|
||||||
| NC score | 0.166363 (rank : 27) | θ value | 3.77169e-06 (rank : 27) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
UBP3_MOUSE
|
||||||
| NC score | 0.163213 (rank : 28) | θ value | 1.53129e-07 (rank : 25) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP20_HUMAN
|
||||||
| NC score | 0.145716 (rank : 29) | θ value | 2.44474e-05 (rank : 29) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.131199 (rank : 30) | θ value | 0.0113563 (rank : 36) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.131038 (rank : 31) | θ value | 0.00298849 (rank : 33) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP13_HUMAN
|
||||||
| NC score | 0.119180 (rank : 32) | θ value | 0.00134147 (rank : 32) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.108704 (rank : 33) | θ value | 2.13673e-09 (rank : 24) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.102930 (rank : 34) | θ value | 0.62314 (rank : 67) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
SELPL_HUMAN
|
||||||
| NC score | 0.102047 (rank : 35) | θ value | 0.00134147 (rank : 31) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.100084 (rank : 36) | θ value | 0.125558 (rank : 46) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.099852 (rank : 37) | θ value | 0.125558 (rank : 45) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP22_HUMAN
|
||||||
| NC score | 0.094847 (rank : 38) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| NC score | 0.093353 (rank : 39) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.076405 (rank : 40) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.075373 (rank : 41) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.072813 (rank : 42) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP21_MOUSE
|
||||||
| NC score | 0.072751 (rank : 43) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP2_MOUSE
|
||||||
| NC score | 0.072676 (rank : 44) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.072349 (rank : 45) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.072221 (rank : 46) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP50_HUMAN
|
||||||
| NC score | 0.072152 (rank : 47) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP21_HUMAN
|
||||||
| NC score | 0.072094 (rank : 48) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP2_HUMAN
|
||||||
| NC score | 0.072050 (rank : 49) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.071181 (rank : 50) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
NUP62_MOUSE
|
||||||
| NC score | 0.071179 (rank : 51) | θ value | 0.00665767 (rank : 35) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.070160 (rank : 52) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP12_HUMAN
|
||||||
| NC score | 0.068266 (rank : 53) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
PME17_HUMAN
|
||||||
| NC score | 0.067651 (rank : 54) | θ value | 0.0736092 (rank : 42) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40967, Q12763, Q14448, Q14817, Q16565 | Gene names | SILV, D12S53E, PMEL17 | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Melanocyte lineage-specific antigen GP100) (Melanoma-associated ME20 antigen) (ME20M/ME20S) (ME20- M/ME20-S) (95 kDa melanocyte-specific secreted glycoprotein). | |||||
|
UBP46_HUMAN
|
||||||
| NC score | 0.067403 (rank : 55) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| NC score | 0.067403 (rank : 56) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.066990 (rank : 57) | θ value | 0.125558 (rank : 44) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
UBPW_MOUSE
|
||||||
| NC score | 0.065652 (rank : 58) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UB17L_HUMAN
|
||||||
| NC score | 0.064334 (rank : 59) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.064282 (rank : 60) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.063922 (rank : 61) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
UBP30_HUMAN
|
||||||
| NC score | 0.063868 (rank : 62) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.063818 (rank : 63) | θ value | 0.00035302 (rank : 30) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.063102 (rank : 64) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.062979 (rank : 65) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP48_HUMAN
|
||||||
| NC score | 0.062255 (rank : 66) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.059538 (rank : 67) | θ value | 0.163984 (rank : 48) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.059233 (rank : 68) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP4_MOUSE
|
||||||
| NC score | 0.058590 (rank : 69) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.058105 (rank : 70) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.058008 (rank : 71) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.057788 (rank : 72) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP15_MOUSE
|
||||||
| NC score | 0.057582 (rank : 73) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP15_HUMAN
|
||||||
| NC score | 0.057547 (rank : 74) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.057323 (rank : 75) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP18_HUMAN
|
||||||
| NC score | 0.056876 (rank : 76) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.056793 (rank : 77) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
PME17_MOUSE
|
||||||
| NC score | 0.056427 (rank : 78) | θ value | 0.0563607 (rank : 41) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.055946 (rank : 79) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.052377 (rank : 80) | θ value | 0.0148317 (rank : 38) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.052016 (rank : 81) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SYCP3_HUMAN
|
||||||
| NC score | 0.051827 (rank : 82) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZU3 | Gene names | SYCP3, SCP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 3 (SCP-3). | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.051774 (rank : 83) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.051319 (rank : 84) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
SNUT2_MOUSE
|
||||||
| NC score | 0.051234 (rank : 85) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
NUP62_HUMAN
|
||||||
| NC score | 0.051122 (rank : 86) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.051063 (rank : 87) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
SNUT2_HUMAN
|
||||||
| NC score | 0.050771 (rank : 88) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
UBP40_HUMAN
|
||||||
| NC score | 0.050072 (rank : 89) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.047433 (rank : 90) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.044188 (rank : 91) | θ value | 0.0563607 (rank : 40) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.043207 (rank : 92) | θ value | 0.279714 (rank : 50) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NFYA_HUMAN
|
||||||
| NC score | 0.042908 (rank : 93) | θ value | 0.279714 (rank : 52) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23511, Q8IXU0 | Gene names | NFYA | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit alpha (Nuclear transcription factor Y subunit A) (NF-YA) (CAAT-box DNA-binding protein subunit A). | |||||
|
SDC3_MOUSE
|
||||||
| NC score | 0.042182 (rank : 94) | θ value | 0.365318 (rank : 54) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64519, Q3UDD9, Q6ZQA4, Q7TQD4 | Gene names | Sdc3, Kiaa0468 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-3 precursor (SYND3). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.042056 (rank : 95) | θ value | 0.163984 (rank : 47) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.041517 (rank : 96) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.041445 (rank : 97) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.041286 (rank : 98) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.040632 (rank : 99) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.040490 (rank : 100) | θ value | 0.0148317 (rank : 37) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.040359 (rank : 101) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.038458 (rank : 102) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
NFRKB_HUMAN
|
||||||
| NC score | 0.037438 (rank : 103) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
K0240_HUMAN
|
||||||
| NC score | 0.037355 (rank : 104) | θ value | 0.125558 (rank : 43) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
SIAL_MOUSE
|
||||||
| NC score | 0.036604 (rank : 105) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61711, Q61363 | Gene names | Ibsp | |||
|
Domain Architecture |

|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.034685 (rank : 106) | θ value | 0.62314 (rank : 68) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
OVGP1_MOUSE
|
||||||
| NC score | 0.032334 (rank : 107) | θ value | 0.00390308 (rank : 34) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62010 | Gene names | Ovgp1, Chit5, Ogp | |||
|
Domain Architecture |

|
|||||
| Description | Oviduct-specific glycoprotein precursor (Oviductal glycoprotein) (Oviductin) (Estrogen-dependent oviduct protein). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.031189 (rank : 108) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.030406 (rank : 109) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.030328 (rank : 110) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
ALG2_MOUSE
|
||||||
| NC score | 0.028630 (rank : 111) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBE8, Q7TN30, Q9CWI6 | Gene names | Alg2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1,3-mannosyltransferase ALG2 (EC 2.4.1.-) (GDP- Man:Man(1)GlcNAc(2)-PP-dolichol mannosyltransferase). | |||||
|
TBP_HUMAN
|
||||||
| NC score | 0.028590 (rank : 112) | θ value | 0.813845 (rank : 71) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20226, Q16845, Q9UC02 | Gene names | TBP, TF2D, TFIID | |||
|
Domain Architecture |

|
|||||
| Description | TATA-box-binding protein (TATA-box factor) (TATA-binding factor) (TATA sequence-binding protein) (Transcription initiation factor TFIID TBP subunit). | |||||
|
A4GCT_HUMAN
|
||||||
| NC score | 0.027562 (rank : 113) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNA3 | Gene names | A4GNT | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1,4-N-acetylglucosaminyltransferase (EC 2.4.1.-) (Alpha4GnT). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.027388 (rank : 114) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
NUPL1_MOUSE
|
||||||
| NC score | 0.027097 (rank : 115) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R332, Q3UJF4, Q8BUA7, Q8BVG7, Q8C0W3 | Gene names | Nupl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin p58/p45 (Nucleoporin-like 1). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.026391 (rank : 116) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.025954 (rank : 117) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SIAL_HUMAN
|
||||||
| NC score | 0.024038 (rank : 118) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |

|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.023458 (rank : 119) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
PORIM_HUMAN
|
||||||
| NC score | 0.022657 (rank : 120) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N131, Q8IWS2, Q96QV2 | Gene names | TMEM123, KCT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123) (Pro-oncosis receptor inducing membrane injury) (Keratinocytes-associated transmembrane protein 3) (KCT-3). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.022150 (rank : 121) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.021661 (rank : 122) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.021473 (rank : 123) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.021364 (rank : 124) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
MA2B2_MOUSE
|
||||||
| NC score | 0.020101 (rank : 125) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54782, Q69ZV1, Q8BH85, Q9DBK2 | Gene names | Man2b2, Kiaa0935 | |||
|
Domain Architecture |

|
|||||
| Description | Epididymis-specific alpha-mannosidase precursor (EC 3.2.1.24) (Mannosidase alpha class 2B member 2). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.020026 (rank : 126) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.019784 (rank : 127) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.019292 (rank : 128) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
T240L_MOUSE
|
||||||
| NC score | 0.018923 (rank : 129) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
ADNP_HUMAN
|
||||||
| NC score | 0.018367 (rank : 130) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H2P0, O94881, Q9UG34 | Gene names | ADNP, KIAA0784 | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.017932 (rank : 131) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.017647 (rank : 132) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.017647 (rank : 133) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
OTU7A_MOUSE
|
||||||
| NC score | 0.017566 (rank : 134) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.017560 (rank : 135) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
ACD_HUMAN
|
||||||
| NC score | 0.017418 (rank : 136) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96AP0, Q562H5, Q9H8F9 | Gene names | ACD, PIP1, PTOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adrenocortical dysplasia protein homolog (POT1 and TIN2-interacting protein). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.017399 (rank : 137) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
CF142_MOUSE
|
||||||
| NC score | 0.017368 (rank : 138) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5FW52, Q9D6X9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf142 homolog. | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.017259 (rank : 139) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
CCG8_MOUSE
|
||||||
| NC score | 0.016904 (rank : 140) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHW2 | Gene names | Cacng8 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-8 subunit (Neuronal voltage- gated calcium channel gamma-8 subunit). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.016347 (rank : 141) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
VASP_MOUSE
|
||||||
| NC score | 0.016053 (rank : 142) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.015772 (rank : 143) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.015203 (rank : 144) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.014652 (rank : 145) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
UBN1_HUMAN
|
||||||
| NC score | 0.013662 (rank : 146) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.013353 (rank : 147) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
RFX1_HUMAN
|
||||||
| NC score | 0.013229 (rank : 148) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
ASH2L_MOUSE
|
||||||
| NC score | 0.011846 (rank : 149) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
ICAL_MOUSE
|
||||||
| NC score | 0.011836 (rank : 150) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.010986 (rank : 151) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CUGB1_MOUSE
|
||||||
| NC score | 0.010825 (rank : 152) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28659, Q9CXE5, Q9EPJ8, Q9JI37 | Gene names | Cugbp1, Brunol2, Cugbp | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (Deadenylation factor EDEN-BP) (Brain protein F41). | |||||
|
CUGB1_HUMAN
|
||||||
| NC score | 0.010813 (rank : 153) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92879, Q9NP83, Q9NR06 | Gene names | CUGBP1, BRUNOL2, CUGBP, NAB50 | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (50 kDa Nuclear polyadenylated RNA-binding protein) (EDEN-BP). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.010529 (rank : 154) | θ value | 0.0330416 (rank : 39) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
DMN_HUMAN
|
||||||
| NC score | 0.010003 (rank : 155) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.009999 (rank : 156) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.009780 (rank : 157) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
CLIC6_MOUSE
|
||||||
| NC score | 0.009683 (rank : 158) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.008676 (rank : 159) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.008567 (rank : 160) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
I17RA_HUMAN
|
||||||
| NC score | 0.008565 (rank : 161) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96F46, O43844 | Gene names | IL17RA, IL17R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-17 receptor A precursor (IL-17 receptor). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.008390 (rank : 162) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
PHF2_HUMAN
|
||||||
| NC score | 0.007971 (rank : 163) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.007773 (rank : 164) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.007682 (rank : 165) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
LRBA_HUMAN
|
||||||
| NC score | 0.007552 (rank : 166) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
UBQL1_MOUSE
|
||||||
| NC score | 0.006741 (rank : 167) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R317, Q80V10, Q8C7T4, Q8C835, Q8K141, Q91VI8, Q9D0Z0, Q9QZM1 | Gene names | Ubqln1, Plic1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-1 (Protein linking IAP with cytoskeleton 1) (PLIC-1). | |||||
|
TRBP2_MOUSE
|
||||||
| NC score | 0.006538 (rank : 168) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.006451 (rank : 169) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.006229 (rank : 170) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
MAGAB_HUMAN
|
||||||
| NC score | 0.005928 (rank : 171) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
PTPR2_HUMAN
|
||||||
| NC score | 0.005505 (rank : 172) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
AUXI_MOUSE
|
||||||
| NC score | 0.005485 (rank : 173) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.004812 (rank : 174) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
CAMKV_MOUSE
|
||||||
| NC score | 0.004165 (rank : 175) | θ value | 0.21417 (rank : 49) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UHL1, Q8VD20 | Gene names | Camkv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
TAOK3_MOUSE
|
||||||
| NC score | 0.003707 (rank : 176) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
SEM4D_MOUSE
|
||||||
| NC score | 0.003509 (rank : 177) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.003405 (rank : 178) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.002721 (rank : 179) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
BMP5_MOUSE
|
||||||
| NC score | 0.001565 (rank : 180) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49003 | Gene names | Bmp5, Bmp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 5 precursor (BMP-5). | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.001353 (rank : 181) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
ZN335_HUMAN
|
||||||
| NC score | -0.000874 (rank : 182) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
LGR5_MOUSE
|
||||||
| NC score | -0.001443 (rank : 183) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1P4 | Gene names | Lgr5, Fex, Gpr49 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 5 precursor (G-protein coupled receptor 49) (Orphan G-protein coupled receptor FEX). | |||||
|
ZNF76_HUMAN
|
||||||
| NC score | -0.002891 (rank : 184) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36508, Q9BQB2 | Gene names | ZNF76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 76. | |||||