Please be patient as the page loads
|
SEM4D_MOUSE
|
||||||
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SEM4D_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995513 (rank : 2) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q92854, Q7Z5S4 | Gene names | SEMA4D, CD100 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Leukocyte activation antigen CD100) (BB18) (A8) (GR3). | |||||
|
SEM4D_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
SEM4C_MOUSE
|
||||||
| θ value | 5.78879e-140 (rank : 3) | NC score | 0.983856 (rank : 3) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q64151 | Gene names | Sema4c, Semacl1, Semai | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor (Semaphorin I) (Sema I) (Semaphorin C-like 1) (M-Sema F). | |||||
|
SEM4C_HUMAN
|
||||||
| θ value | 9.56396e-135 (rank : 4) | NC score | 0.980893 (rank : 4) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9C0C4, Q7Z5X0 | Gene names | SEMA4C, KIAA1739 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor. | |||||
|
SEM4B_MOUSE
|
||||||
| θ value | 5.08302e-128 (rank : 5) | NC score | 0.978339 (rank : 8) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SEM4B_HUMAN
|
||||||
| θ value | 2.52267e-127 (rank : 6) | NC score | 0.979225 (rank : 5) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
SEM4G_HUMAN
|
||||||
| θ value | 2.61662e-116 (rank : 7) | NC score | 0.977957 (rank : 9) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4G_MOUSE
|
||||||
| θ value | 4.4633e-116 (rank : 8) | NC score | 0.977474 (rank : 10) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4A_HUMAN
|
||||||
| θ value | 5.84277e-108 (rank : 9) | NC score | 0.976179 (rank : 11) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4F_HUMAN
|
||||||
| θ value | 3.20603e-106 (rank : 10) | NC score | 0.978777 (rank : 7) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95754, Q9NS35 | Gene names | SEMA4F, SEMAM, SEMAW | |||
|
Domain Architecture |
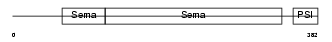
|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W) (Semaphorin M) (Sema M). | |||||
|
SEM4F_MOUSE
|
||||||
| θ value | 1.03134e-104 (rank : 11) | NC score | 0.979155 (rank : 6) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z123, Q9R1Y1 | Gene names | Sema4f, Semaw | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W). | |||||
|
SEM4A_MOUSE
|
||||||
| θ value | 2.37764e-101 (rank : 12) | NC score | 0.975070 (rank : 12) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62178 | Gene names | Sema4a, Semab, SemB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM3A_HUMAN
|
||||||
| θ value | 3.10529e-101 (rank : 13) | NC score | 0.965419 (rank : 16) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14563 | Gene names | SEMA3A | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III). | |||||
|
SEM3A_MOUSE
|
||||||
| θ value | 5.29683e-101 (rank : 14) | NC score | 0.965437 (rank : 15) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O08665, Q5BL08, Q62180, Q62215 | Gene names | Sema3a, Semad, SemD | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III) (Semaphorin D) (Sema D). | |||||
|
SEM3C_HUMAN
|
||||||
| θ value | 4.19693e-98 (rank : 15) | NC score | 0.964246 (rank : 18) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99985 | Gene names | SEMA3C, SEMAE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3C_MOUSE
|
||||||
| θ value | 1.59483e-97 (rank : 16) | NC score | 0.964600 (rank : 17) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62181 | Gene names | Sema3c, Semae, SemE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3B_HUMAN
|
||||||
| θ value | 1.03374e-96 (rank : 17) | NC score | 0.967408 (rank : 13) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||
|
SEM3D_HUMAN
|
||||||
| θ value | 2.38316e-93 (rank : 18) | NC score | 0.962942 (rank : 19) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |
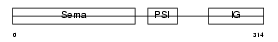
|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3D_MOUSE
|
||||||
| θ value | 2.01746e-92 (rank : 19) | NC score | 0.962044 (rank : 23) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3B_MOUSE
|
||||||
| θ value | 1.03614e-88 (rank : 20) | NC score | 0.967120 (rank : 14) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q62177 | Gene names | Sema3b, Sema, Semaa | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin A) (Sema A). | |||||
|
SEM3E_HUMAN
|
||||||
| θ value | 2.30828e-88 (rank : 21) | NC score | 0.962926 (rank : 20) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O15041, Q75M94, Q75M97 | Gene names | SEMA3E, KIAA0331 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3E precursor. | |||||
|
SEM3E_MOUSE
|
||||||
| θ value | 2.5521e-87 (rank : 22) | NC score | 0.962420 (rank : 22) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70275, O09078, O09079 | Gene names | Sema3e, Semah, Semh | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3E precursor (Semaphorin H) (Sema H). | |||||
|
SEM3F_MOUSE
|
||||||
| θ value | 1.2666e-86 (rank : 23) | NC score | 0.962795 (rank : 21) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88632, O88633 | Gene names | Sema3f | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV). | |||||
|
SEM3F_HUMAN
|
||||||
| θ value | 2.16048e-86 (rank : 24) | NC score | 0.961798 (rank : 24) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13275, Q13274, Q13372, Q15704 | Gene names | SEMA3F | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV) (Sema III/F). | |||||
|
SEM6D_MOUSE
|
||||||
| θ value | 2.02684e-76 (rank : 25) | NC score | 0.942987 (rank : 30) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6D_HUMAN
|
||||||
| θ value | 5.8972e-76 (rank : 26) | NC score | 0.942048 (rank : 31) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6A_HUMAN
|
||||||
| θ value | 1.04096e-72 (rank : 27) | NC score | 0.944617 (rank : 27) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM6A_MOUSE
|
||||||
| θ value | 6.7473e-72 (rank : 28) | NC score | 0.943886 (rank : 28) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
SEM5B_MOUSE
|
||||||
| θ value | 1.71981e-67 (rank : 29) | NC score | 0.793679 (rank : 37) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q60519 | Gene names | Sema5b, Semag, SemG | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor (Semaphorin G) (Sema G). | |||||
|
SEM5B_HUMAN
|
||||||
| θ value | 1.45591e-66 (rank : 30) | NC score | 0.797194 (rank : 35) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9P283, Q6UY12 | Gene names | SEMA5B, KIAA1445 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor. | |||||
|
SEM5A_HUMAN
|
||||||
| θ value | 1.2325e-65 (rank : 31) | NC score | 0.791774 (rank : 38) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13591, O60408, Q1RLL9 | Gene names | SEMA5A, SEMAF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM6C_HUMAN
|
||||||
| θ value | 8.83269e-64 (rank : 32) | NC score | 0.943645 (rank : 29) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM5A_MOUSE
|
||||||
| θ value | 3.35642e-63 (rank : 33) | NC score | 0.796480 (rank : 36) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM6B_MOUSE
|
||||||
| θ value | 6.32992e-62 (rank : 34) | NC score | 0.937344 (rank : 33) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
SEM6B_HUMAN
|
||||||
| θ value | 6.99855e-61 (rank : 35) | NC score | 0.939225 (rank : 32) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 2.65945e-60 (rank : 36) | NC score | 0.928730 (rank : 34) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM7A_MOUSE
|
||||||
| θ value | 7.75577e-52 (rank : 37) | NC score | 0.947959 (rank : 25) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QUR8, O88371 | Gene names | Sema7a, Cd108, Semal, Semk1 | |||
|
Domain Architecture |
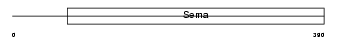
|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (CD108 antigen) (CDw108). | |||||
|
SEM7A_HUMAN
|
||||||
| θ value | 3.84914e-51 (rank : 38) | NC score | 0.947274 (rank : 26) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75326 | Gene names | SEMA7A, CD108, SEMAL | |||
|
Domain Architecture |
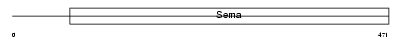
|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (John-Milton-Hargen human blood group Ag) (JMH blood group antigen) (CD108 antigen) (CDw108). | |||||
|
PLXA1_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 39) | NC score | 0.333393 (rank : 46) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXA1_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 40) | NC score | 0.325279 (rank : 48) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
PLXC1_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 41) | NC score | 0.362207 (rank : 41) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXC1_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 42) | NC score | 0.370901 (rank : 40) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXA3_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 43) | NC score | 0.326620 (rank : 47) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 44) | NC score | 0.323332 (rank : 49) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
PLXA2_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 45) | NC score | 0.338755 (rank : 44) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70207, Q6NVE6, Q6PHN4, Q80TZ7, Q8R1I4 | Gene names | Plxna2, Kiaa0463 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Plexin-2) (Plex 2). | |||||
|
PLXA2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 46) | NC score | 0.338525 (rank : 45) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O75051, Q6UX61, Q96GN9, Q9BRL1, Q9UIW1 | Gene names | PLXNA2, KIAA0463, OCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Semaphorin receptor OCT). | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 47) | NC score | 0.320984 (rank : 50) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PLXD1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 48) | NC score | 0.291901 (rank : 53) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
PLXB3_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 49) | NC score | 0.296054 (rank : 52) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
NGL1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 50) | NC score | 0.023543 (rank : 97) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 512 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8C031, Q8BGH8 | Gene names | Lrrc4c, Ngl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-G1 ligand precursor (NGL-1) (Leucine-rich repeat-containing protein 4C). | |||||
|
NGL1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 51) | NC score | 0.023565 (rank : 96) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCJ2, Q7L0N3 | Gene names | LRRC4C, KIAA1580, NGL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-G1 ligand precursor (NGL-1) (Leucine-rich repeat-containing protein 4C). | |||||
|
PLXA4_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 52) | NC score | 0.349385 (rank : 43) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXB3_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 53) | NC score | 0.296711 (rank : 51) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
PLXA4_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 54) | NC score | 0.350220 (rank : 42) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
NCAM2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 55) | NC score | 0.035822 (rank : 75) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15394 | Gene names | NCAM2, NCAM21 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 2 precursor (N-CAM 2). | |||||
|
ATRN_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 56) | NC score | 0.030073 (rank : 84) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
NRCAM_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 57) | NC score | 0.029670 (rank : 86) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92823, O15051, O15179, Q9UHI3, Q9UHI4 | Gene names | NRCAM, KIAA0343 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal cell adhesion molecule precursor (Nr-CAM) (NgCAM-related cell adhesion molecule) (Ng-CAM-related) (hBravo). | |||||
|
PLXB2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 58) | NC score | 0.371871 (rank : 39) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
ATRN_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 59) | NC score | 0.028629 (rank : 87) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
NCAM2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 60) | NC score | 0.034827 (rank : 77) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35136, O35962 | Gene names | Ncam2, Ocam, Rncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 2 precursor (N-CAM 2) (RB-8 neural cell adhesion molecule) (R4B12). | |||||
|
BOC_MOUSE
|
||||||
| θ value | 0.125558 (rank : 61) | NC score | 0.033222 (rank : 80) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6AZB0, Q6KAM5, Q6P5H3, Q7TMJ3, Q8CE73, Q8CE91, Q8R377, Q923W7 | Gene names | Boc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
NEO1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 62) | NC score | 0.034111 (rank : 79) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92859, O00340 | Gene names | NEO1, NGN | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
DCC_HUMAN
|
||||||
| θ value | 0.163984 (rank : 63) | NC score | 0.037654 (rank : 72) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 0.163984 (rank : 64) | NC score | 0.019157 (rank : 104) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 65) | NC score | 0.015274 (rank : 112) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 0.21417 (rank : 66) | NC score | 0.035056 (rank : 76) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
X3CL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 67) | NC score | 0.019207 (rank : 103) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78423, O00672 | Gene names | CX3CL1, A-152E5.2, FKN, NTT, SCYD1 | |||
|
Domain Architecture |
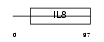
|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
NEO1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 68) | NC score | 0.034563 (rank : 78) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 69) | NC score | 0.027860 (rank : 90) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.365318 (rank : 70) | NC score | 0.015613 (rank : 110) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ROBO3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 71) | NC score | 0.031576 (rank : 82) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.365318 (rank : 72) | NC score | 0.019579 (rank : 101) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CENG1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 73) | NC score | 0.014090 (rank : 115) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
ROR1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 74) | NC score | 0.002519 (rank : 146) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z139 | Gene names | Ror1, Ntrkr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR1 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 1) (mROR1). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.005759 (rank : 134) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NRCAM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.029678 (rank : 85) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q810U4, Q80U33, Q8BLG8, Q8BX92, Q8BYJ8 | Gene names | Nrcam, Kiaa0343 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal cell adhesion molecule precursor (Nr-CAM) (NgCAM-related cell adhesion molecule) (Ng-CAM-related) (mBravo). | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 77) | NC score | 0.017668 (rank : 105) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
SUHW4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.000784 (rank : 153) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.007066 (rank : 128) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 80) | NC score | 0.009289 (rank : 122) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
PXDC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 81) | NC score | 0.022709 (rank : 98) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91ZV7, Q8BM20, Q9CWV5 | Gene names | Plxdc1, Tem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7). | |||||
|
ROBO3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.033007 (rank : 81) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.014610 (rank : 113) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
YETS2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.012323 (rank : 117) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
ESAM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.022586 (rank : 99) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q925F2 | Gene names | Esam, Esam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 1.38821 (rank : 86) | NC score | 0.022205 (rank : 100) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
BOC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.036678 (rank : 74) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
CNTN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.027232 (rank : 92) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12860, Q12861, Q14030, Q8N466 | Gene names | CNTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3) (Glycoprotein gp135). | |||||
|
CNTN4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.048996 (rank : 70) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IWV2, Q8IX14, Q8TC35 | Gene names | CNTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
FMT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.016207 (rank : 107) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D799, Q8VE89 | Gene names | Mtfmt, Fmt | |||
|
Domain Architecture |
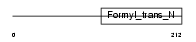
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
POGZ_MOUSE
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.000319 (rank : 155) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
DRAP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.015388 (rank : 111) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14919, Q13448 | Gene names | DRAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
LSAMP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.027357 (rank : 91) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BLK3 | Gene names | Lsamp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbic system-associated membrane protein precursor (LSAMP). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.003999 (rank : 137) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NEIL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.011745 (rank : 118) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96FI4, Q6ZRA7, Q86XW7, Q9H6C3 | Gene names | NEIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1) (FPG1). | |||||
|
TEAD3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.005604 (rank : 135) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70210, O08516, O70623, P70209 | Gene names | Tead3, Tcf13r2, Tef5 | |||
|
Domain Architecture |
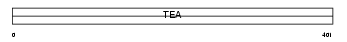
|
|||||
| Description | Transcriptional enhancer factor TEF-5 (TEA domain family member 3) (TEAD-3) (ETF-related factor 1) (ETFR-1) (DTEF-1). | |||||
|
CNTN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.026748 (rank : 93) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12960, Q6NXV7, Q8BR42, Q8C6A0 | Gene names | Cntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3). | |||||
|
LSAMP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.026653 (rank : 94) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13449, Q8IV49 | Gene names | LSAMP, LAMP | |||
|
Domain Architecture |
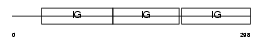
|
|||||
| Description | Limbic system-associated membrane protein precursor (LSAMP). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.011017 (rank : 121) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.019465 (rank : 102) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
ROBO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.028503 (rank : 88) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
ROBO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.028398 (rank : 89) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.008705 (rank : 124) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.002535 (rank : 145) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TRPV1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.003192 (rank : 143) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
FGFR3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.003119 (rank : 144) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61851, Q61564, Q63834 | Gene names | Fgfr3, Mfr3, Sam3 | |||
|
Domain Architecture |

|
|||||
| Description | Fibroblast growth factor receptor 3 precursor (EC 2.7.10.1) (FGFR-3) (Heparin-binding growth factor receptor) (CD333 antigen). | |||||
|
FMT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.014127 (rank : 114) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DP5 | Gene names | MTFMT, FMT, FMT1 | |||
|
Domain Architecture |
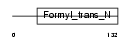
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
GP151_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.002475 (rank : 147) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDV0, Q86SN8, Q8NGV2 | Gene names | GPR151, PGR7 | |||
|
Domain Architecture |
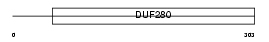
|
|||||
| Description | Probable G-protein coupled receptor 151 (G-protein coupled receptor PGR7) (GPCR-2037). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.003806 (rank : 140) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
POGZ_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | -0.000114 (rank : 156) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z3K3, O75049, Q8TDZ7, Q9Y4X7 | Gene names | POGZ, KIAA0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
RANB9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.016448 (rank : 106) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
SCG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.006621 (rank : 129) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
SNCAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.002417 (rank : 148) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.005825 (rank : 133) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
HDAC6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.003509 (rank : 141) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
JAM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.037206 (rank : 73) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y624 | Gene names | F11R, JAM1, JCAM | |||
|
Domain Architecture |
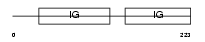
|
|||||
| Description | Junctional adhesion molecule A precursor (JAM-A) (Junctional adhesion molecule 1) (JAM-1) (Platelet adhesion molecule 1) (PAM-1) (Platelet F11 receptor) (CD321 antigen). | |||||
|
LRC4B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.015975 (rank : 109) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NT99, Q3ZCQ4, Q58F20 | Gene names | LRRC4B, LRIG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 4B precursor. | |||||
|
LRC4B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.016121 (rank : 108) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P0C192 | Gene names | Lrrc4b, Lrig4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 4B precursor. | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.006251 (rank : 130) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
UTP20_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.005957 (rank : 131) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5XG71, Q80V22, Q8BXH9, Q8CHL6, Q99K11 | Gene names | Utp20, Drim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein). | |||||
|
VGFR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.003963 (rank : 138) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35968, O60723, Q14178 | Gene names | KDR, FLK1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 2 precursor (EC 2.7.10.1) (VEGFR-2) (Kinase insert domain receptor) (Protein-tyrosine kinase receptor Flk-1) (CD309 antigen). | |||||
|
ZDHC8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.002315 (rank : 149) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.001964 (rank : 150) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CJ026_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.005841 (rank : 132) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGW2, Q3TF48, Q3U3M2, Q6PD43, Q8R0W8 | Gene names | D19Wsu162e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf26 homolog. | |||||
|
DAF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.001647 (rank : 151) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61476 | Gene names | Cd55b, Daf2 | |||
|
Domain Architecture |
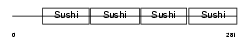
|
|||||
| Description | Complement decay-accelerating factor transmembrane isoform precursor (DAF-TM) (CD55 antigen). | |||||
|
DRAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.011536 (rank : 120) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D6N5 | Gene names | Drap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
GPR83_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | -0.005654 (rank : 158) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 890 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYM4, Q9P1Y8 | Gene names | GPR83, GPR72, KIAA1540 | |||
|
Domain Architecture |
|
|||||
| Description | Probable G-protein coupled receptor 83 precursor (G-protein coupled receptor 72). | |||||
|
HMCN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.065906 (rank : 60) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.009079 (rank : 123) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
PAPOA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.007788 (rank : 125) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
TROP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.001638 (rank : 152) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.003266 (rank : 142) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
ABI2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.000498 (rank : 154) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
ATE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.012373 (rank : 116) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2A5, Q9Z2A4 | Gene names | Ate1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA--protein transferase 1 (EC 2.3.2.8) (R-transferase 1) (Arginyltransferase 1) (Arginine-tRNA--protein transferase 1). | |||||
|
CDON_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.030491 (rank : 83) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q32MD9, O88971 | Gene names | Cdon, Cdo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | -0.000277 (rank : 157) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CK024_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.007731 (rank : 126) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
IGSF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.011744 (rank : 119) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q93033, Q15856 | Gene names | IGSF2, CD101, EWI101, V7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin superfamily member 2 precursor (Glu-Trp-Ile EWI motif- containing protein 101) (EWI-101) (Cell surface glycoprotein V7) (CD101 antigen). | |||||
|
OBSL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.044507 (rank : 71) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75147, Q96IW3 | Gene names | OBSL1, KIAA0657 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin-like protein 1 precursor. | |||||
|
PHAR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.024110 (rank : 95) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.007085 (rank : 127) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.004724 (rank : 136) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.003850 (rank : 139) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
BAI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.052705 (rank : 67) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BAI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.052509 (rank : 68) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
CILP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.052244 (rank : 69) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IUL8, Q6NV88, Q8N4A6, Q8WV21 | Gene names | CILP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 2 precursor (CILP-2) [Contains: Cartilage intermediate layer protein 2 C1; Cartilage intermediate layer protein 2 C2]. | |||||
|
PROP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.092270 (rank : 54) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27918, O15134, O15135, O15136, O75826 | Gene names | CFP, PFC | |||
|
Domain Architecture |
|
|||||
| Description | Properdin precursor (Factor P). | |||||
|
PROP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.090437 (rank : 55) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
SPON1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.066790 (rank : 59) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCB6, O94862, Q8NCD7, Q8WUR5 | Gene names | SPON1, KIAA0762, VSGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin) (Vascular smooth muscle cell growth- promoting factor). | |||||
|
SPON1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.068353 (rank : 58) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VCC9, Q8K2Q8 | Gene names | Spon1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.053387 (rank : 66) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
UNC5A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.061464 (rank : 64) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K1S4, Q6PEF7, Q80T71 | Gene names | Unc5a, Kiaa1976, Unc5h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5A precursor (Unc-5 homolog A) (Unc-5 homolog 1). | |||||
|
UNC5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.064998 (rank : 61) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
UNC5B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.064565 (rank : 62) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1S3, Q3U4F2, Q6PFH0, Q80Y85, Q9D398 | Gene names | Unc5b, Unc5h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2). | |||||
|
UNC5C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.061191 (rank : 65) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95185, Q8IUT0 | Gene names | UNC5C, UNC5H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3). | |||||
|
UNC5C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.061900 (rank : 63) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08747, Q8CD16 | Gene names | Unc5c, Rcm, Unc5h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3) (Rostral cerebellar malformation protein). | |||||
|
UNC5D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.074954 (rank : 57) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6UXZ4, Q8WYP7 | Gene names | UNC5D, KIAA1777, UNC5H4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
UNC5D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.075177 (rank : 56) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K1S2 | Gene names | Unc5d, Unc5h4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
SEM4D_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
SEM4D_HUMAN
|
||||||
| NC score | 0.995513 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q92854, Q7Z5S4 | Gene names | SEMA4D, CD100 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Leukocyte activation antigen CD100) (BB18) (A8) (GR3). | |||||
|
SEM4C_MOUSE
|
||||||
| NC score | 0.983856 (rank : 3) | θ value | 5.78879e-140 (rank : 3) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q64151 | Gene names | Sema4c, Semacl1, Semai | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor (Semaphorin I) (Sema I) (Semaphorin C-like 1) (M-Sema F). | |||||
|
SEM4C_HUMAN
|
||||||
| NC score | 0.980893 (rank : 4) | θ value | 9.56396e-135 (rank : 4) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9C0C4, Q7Z5X0 | Gene names | SEMA4C, KIAA1739 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor. | |||||
|
SEM4B_HUMAN
|
||||||
| NC score | 0.979225 (rank : 5) | θ value | 2.52267e-127 (rank : 6) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
SEM4F_MOUSE
|
||||||
| NC score | 0.979155 (rank : 6) | θ value | 1.03134e-104 (rank : 11) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z123, Q9R1Y1 | Gene names | Sema4f, Semaw | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W). | |||||
|
SEM4F_HUMAN
|
||||||
| NC score | 0.978777 (rank : 7) | θ value | 3.20603e-106 (rank : 10) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95754, Q9NS35 | Gene names | SEMA4F, SEMAM, SEMAW | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W) (Semaphorin M) (Sema M). | |||||
|
SEM4B_MOUSE
|
||||||
| NC score | 0.978339 (rank : 8) | θ value | 5.08302e-128 (rank : 5) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SEM4G_HUMAN
|
||||||
| NC score | 0.977957 (rank : 9) | θ value | 2.61662e-116 (rank : 7) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4G_MOUSE
|
||||||
| NC score | 0.977474 (rank : 10) | θ value | 4.4633e-116 (rank : 8) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4A_HUMAN
|
||||||
| NC score | 0.976179 (rank : 11) | θ value | 5.84277e-108 (rank : 9) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4A_MOUSE
|
||||||
| NC score | 0.975070 (rank : 12) | θ value | 2.37764e-101 (rank : 12) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62178 | Gene names | Sema4a, Semab, SemB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM3B_HUMAN
|
||||||
| NC score | 0.967408 (rank : 13) | θ value | 1.03374e-96 (rank : 17) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||
|
SEM3B_MOUSE
|
||||||
| NC score | 0.967120 (rank : 14) | θ value | 1.03614e-88 (rank : 20) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q62177 | Gene names | Sema3b, Sema, Semaa | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin A) (Sema A). | |||||
|
SEM3A_MOUSE
|
||||||
| NC score | 0.965437 (rank : 15) | θ value | 5.29683e-101 (rank : 14) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O08665, Q5BL08, Q62180, Q62215 | Gene names | Sema3a, Semad, SemD | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III) (Semaphorin D) (Sema D). | |||||
|
SEM3A_HUMAN
|
||||||
| NC score | 0.965419 (rank : 16) | θ value | 3.10529e-101 (rank : 13) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14563 | Gene names | SEMA3A | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III). | |||||
|
SEM3C_MOUSE
|
||||||
| NC score | 0.964600 (rank : 17) | θ value | 1.59483e-97 (rank : 16) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62181 | Gene names | Sema3c, Semae, SemE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3C_HUMAN
|
||||||
| NC score | 0.964246 (rank : 18) | θ value | 4.19693e-98 (rank : 15) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99985 | Gene names | SEMA3C, SEMAE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3D_HUMAN
|
||||||
| NC score | 0.962942 (rank : 19) | θ value | 2.38316e-93 (rank : 18) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3E_HUMAN
|
||||||
| NC score | 0.962926 (rank : 20) | θ value | 2.30828e-88 (rank : 21) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O15041, Q75M94, Q75M97 | Gene names | SEMA3E, KIAA0331 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3E precursor. | |||||
|
SEM3F_MOUSE
|
||||||
| NC score | 0.962795 (rank : 21) | θ value | 1.2666e-86 (rank : 23) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88632, O88633 | Gene names | Sema3f | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV). | |||||
|
SEM3E_MOUSE
|
||||||
| NC score | 0.962420 (rank : 22) | θ value | 2.5521e-87 (rank : 22) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70275, O09078, O09079 | Gene names | Sema3e, Semah, Semh | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3E precursor (Semaphorin H) (Sema H). | |||||
|
SEM3D_MOUSE
|
||||||
| NC score | 0.962044 (rank : 23) | θ value | 2.01746e-92 (rank : 19) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3F_HUMAN
|
||||||
| NC score | 0.961798 (rank : 24) | θ value | 2.16048e-86 (rank : 24) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13275, Q13274, Q13372, Q15704 | Gene names | SEMA3F | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV) (Sema III/F). | |||||
|
SEM7A_MOUSE
|
||||||
| NC score | 0.947959 (rank : 25) | θ value | 7.75577e-52 (rank : 37) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QUR8, O88371 | Gene names | Sema7a, Cd108, Semal, Semk1 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (CD108 antigen) (CDw108). | |||||
|
SEM7A_HUMAN
|
||||||
| NC score | 0.947274 (rank : 26) | θ value | 3.84914e-51 (rank : 38) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75326 | Gene names | SEMA7A, CD108, SEMAL | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (John-Milton-Hargen human blood group Ag) (JMH blood group antigen) (CD108 antigen) (CDw108). | |||||
|
SEM6A_HUMAN
|
||||||
| NC score | 0.944617 (rank : 27) | θ value | 1.04096e-72 (rank : 27) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM6A_MOUSE
|
||||||
| NC score | 0.943886 (rank : 28) | θ value | 6.7473e-72 (rank : 28) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
SEM6C_HUMAN
|
||||||
| NC score | 0.943645 (rank : 29) | θ value | 8.83269e-64 (rank : 32) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM6D_MOUSE
|
||||||
| NC score | 0.942987 (rank : 30) | θ value | 2.02684e-76 (rank : 25) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6D_HUMAN
|
||||||
| NC score | 0.942048 (rank : 31) | θ value | 5.8972e-76 (rank : 26) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6B_HUMAN
|
||||||
| NC score | 0.939225 (rank : 32) | θ value | 6.99855e-61 (rank : 35) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SEM6B_MOUSE
|
||||||
| NC score | 0.937344 (rank : 33) | θ value | 6.32992e-62 (rank : 34) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.928730 (rank : 34) | θ value | 2.65945e-60 (rank : 36) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM5B_HUMAN
|
||||||
| NC score | 0.797194 (rank : 35) | θ value | 1.45591e-66 (rank : 30) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9P283, Q6UY12 | Gene names | SEMA5B, KIAA1445 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor. | |||||
|
SEM5A_MOUSE
|
||||||
| NC score | 0.796480 (rank : 36) | θ value | 3.35642e-63 (rank : 33) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM5B_MOUSE
|
||||||
| NC score | 0.793679 (rank : 37) | θ value | 1.71981e-67 (rank : 29) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q60519 | Gene names | Sema5b, Semag, SemG | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor (Semaphorin G) (Sema G). | |||||
|
SEM5A_HUMAN
|
||||||
| NC score | 0.791774 (rank : 38) | θ value | 1.2325e-65 (rank : 31) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13591, O60408, Q1RLL9 | Gene names | SEMA5A, SEMAF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
PLXB2_HUMAN
|
||||||
| NC score | 0.371871 (rank : 39) | θ value | 0.0431538 (rank : 58) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
PLXC1_MOUSE
|
||||||
| NC score | 0.370901 (rank : 40) | θ value | 4.92598e-06 (rank : 42) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXC1_HUMAN
|
||||||
| NC score | 0.362207 (rank : 41) | θ value | 1.29631e-06 (rank : 41) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXA4_HUMAN
|
||||||
| NC score | 0.350220 (rank : 42) | θ value | 0.00665767 (rank : 54) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA4_MOUSE
|
||||||
| NC score | 0.349385 (rank : 43) | θ value | 0.00298849 (rank : 52) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA2_MOUSE
|
||||||
| NC score | 0.338755 (rank : 44) | θ value | 5.44631e-05 (rank : 45) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70207, Q6NVE6, Q6PHN4, Q80TZ7, Q8R1I4 | Gene names | Plxna2, Kiaa0463 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Plexin-2) (Plex 2). | |||||
|
PLXA2_HUMAN
|
||||||
| NC score | 0.338525 (rank : 45) | θ value | 0.00020696 (rank : 46) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O75051, Q6UX61, Q96GN9, Q9BRL1, Q9UIW1 | Gene names | PLXNA2, KIAA0463, OCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Semaphorin receptor OCT). | |||||
|
PLXA1_MOUSE
|
||||||
| NC score | 0.333393 (rank : 46) | θ value | 5.26297e-08 (rank : 39) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXA3_HUMAN
|
||||||
| NC score | 0.326620 (rank : 47) | θ value | 6.43352e-06 (rank : 43) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
PLXA1_HUMAN
|
||||||
| NC score | 0.325279 (rank : 48) | θ value | 1.17247e-07 (rank : 40) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.323332 (rank : 49) | θ value | 3.19293e-05 (rank : 44) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.320984 (rank : 50) | θ value | 0.00020696 (rank : 47) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PLXB3_MOUSE
|
||||||
| NC score | 0.296711 (rank : 51) | θ value | 0.00390308 (rank : 53) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
PLXB3_HUMAN
|
||||||
| NC score | 0.296054 (rank : 52) | θ value | 0.00035302 (rank : 49) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
PLXD1_HUMAN
|
||||||
| NC score | 0.291901 (rank : 53) | θ value | 0.00020696 (rank : 48) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
PROP_HUMAN
|
||||||
| NC score | 0.092270 (rank : 54) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27918, O15134, O15135, O15136, O75826 | Gene names | CFP, PFC | |||
|
Domain Architecture |

|
|||||
| Description | Properdin precursor (Factor P). | |||||
|
PROP_MOUSE
|
||||||
| NC score | 0.090437 (rank : 55) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
UNC5D_MOUSE
|
||||||
| NC score | 0.075177 (rank : 56) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K1S2 | Gene names | Unc5d, Unc5h4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
UNC5D_HUMAN
|
||||||
| NC score | 0.074954 (rank : 57) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6UXZ4, Q8WYP7 | Gene names | UNC5D, KIAA1777, UNC5H4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
SPON1_MOUSE
|
||||||
| NC score | 0.068353 (rank : 58) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VCC9, Q8K2Q8 | Gene names | Spon1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin). | |||||
|
SPON1_HUMAN
|
||||||
| NC score | 0.066790 (rank : 59) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCB6, O94862, Q8NCD7, Q8WUR5 | Gene names | SPON1, KIAA0762, VSGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin) (Vascular smooth muscle cell growth- promoting factor). | |||||
|
HMCN1_HUMAN
|
||||||
| NC score | 0.065906 (rank : 60) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
UNC5B_HUMAN
|
||||||
| NC score | 0.064998 (rank : 61) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
UNC5B_MOUSE
|
||||||
| NC score | 0.064565 (rank : 62) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1S3, Q3U4F2, Q6PFH0, Q80Y85, Q9D398 | Gene names | Unc5b, Unc5h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2). | |||||
|
UNC5C_MOUSE
|
||||||
| NC score | 0.061900 (rank : 63) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08747, Q8CD16 | Gene names | Unc5c, Rcm, Unc5h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3) (Rostral cerebellar malformation protein). | |||||
|
UNC5A_MOUSE
|
||||||
| NC score | 0.061464 (rank : 64) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K1S4, Q6PEF7, Q80T71 | Gene names | Unc5a, Kiaa1976, Unc5h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5A precursor (Unc-5 homolog A) (Unc-5 homolog 1). | |||||
|
UNC5C_HUMAN
|
||||||
| NC score | 0.061191 (rank : 65) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95185, Q8IUT0 | Gene names | UNC5C, UNC5H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.053387 (rank : 66) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.052705 (rank : 67) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BAI1_MOUSE
|
||||||
| NC score | 0.052509 (rank : 68) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
CILP2_HUMAN
|
||||||
| NC score | 0.052244 (rank : 69) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IUL8, Q6NV88, Q8N4A6, Q8WV21 | Gene names | CILP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 2 precursor (CILP-2) [Contains: Cartilage intermediate layer protein 2 C1; Cartilage intermediate layer protein 2 C2]. | |||||
|
CNTN4_HUMAN
|
||||||
| NC score | 0.048996 (rank : 70) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IWV2, Q8IX14, Q8TC35 | Gene names | CNTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
OBSL1_HUMAN
|
||||||
| NC score | 0.044507 (rank : 71) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75147, Q96IW3 | Gene names | OBSL1, KIAA0657 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin-like protein 1 precursor. | |||||
|
DCC_HUMAN
|
||||||
| NC score | 0.037654 (rank : 72) | θ value | 0.163984 (rank : 63) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
JAM1_HUMAN
|
||||||
| NC score | 0.037206 (rank : 73) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y624 | Gene names | F11R, JAM1, JCAM | |||
|
Domain Architecture |

|
|||||
| Description | Junctional adhesion molecule A precursor (JAM-A) (Junctional adhesion molecule 1) (JAM-1) (Platelet adhesion molecule 1) (PAM-1) (Platelet F11 receptor) (CD321 antigen). | |||||
|
BOC_HUMAN
|
||||||
| NC score | 0.036678 (rank : 74) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
NCAM2_HUMAN
|
||||||
| NC score | 0.035822 (rank : 75) | θ value | 0.0193708 (rank : 55) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15394 | Gene names | NCAM2, NCAM21 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 2 precursor (N-CAM 2). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.035056 (rank : 76) | θ value | 0.21417 (rank : 66) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
NCAM2_MOUSE
|
||||||
| NC score | 0.034827 (rank : 77) | θ value | 0.0736092 (rank : 60) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35136, O35962 | Gene names | Ncam2, Ocam, Rncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 2 precursor (N-CAM 2) (RB-8 neural cell adhesion molecule) (R4B12). | |||||
|
NEO1_MOUSE
|
||||||
| NC score | 0.034563 (rank : 78) | θ value | 0.279714 (rank : 68) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
NEO1_HUMAN
|
||||||
| NC score | 0.034111 (rank : 79) | θ value | 0.125558 (rank : 62) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92859, O00340 | Gene names | NEO1, NGN | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
BOC_MOUSE
|
||||||
| NC score | 0.033222 (rank : 80) | θ value | 0.125558 (rank : 61) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6AZB0, Q6KAM5, Q6P5H3, Q7TMJ3, Q8CE73, Q8CE91, Q8R377, Q923W7 | Gene names | Boc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
ROBO3_HUMAN
|
||||||
| NC score | 0.033007 (rank : 81) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
ROBO3_MOUSE
|
||||||
| NC score | 0.031576 (rank : 82) | θ value | 0.365318 (rank : 71) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
CDON_MOUSE
|
||||||
| NC score | 0.030491 (rank : 83) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q32MD9, O88971 | Gene names | Cdon, Cdo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.030073 (rank : 84) | θ value | 0.0330416 (rank : 56) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
NRCAM_MOUSE
|
||||||
| NC score | 0.029678 (rank : 85) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q810U4, Q80U33, Q8BLG8, Q8BX92, Q8BYJ8 | Gene names | Nrcam, Kiaa0343 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal cell adhesion molecule precursor (Nr-CAM) (NgCAM-related cell adhesion molecule) (Ng-CAM-related) (mBravo). | |||||
|
NRCAM_HUMAN
|
||||||
| NC score | 0.029670 (rank : 86) | θ value | 0.0330416 (rank : 57) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92823, O15051, O15179, Q9UHI3, Q9UHI4 | Gene names | NRCAM, KIAA0343 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal cell adhesion molecule precursor (Nr-CAM) (NgCAM-related cell adhesion molecule) (Ng-CAM-related) (hBravo). | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.028629 (rank : 87) | θ value | 0.0736092 (rank : 59) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
ROBO1_HUMAN
|
||||||
| NC score | 0.028503 (rank : 88) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
ROBO1_MOUSE
|
||||||
| NC score | 0.028398 (rank : 89) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.027860 (rank : 90) | θ value | 0.279714 (rank : 69) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
LSAMP_MOUSE
|
||||||
| NC score | 0.027357 (rank : 91) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BLK3 | Gene names | Lsamp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbic system-associated membrane protein precursor (LSAMP). | |||||
|
CNTN1_HUMAN
|
||||||
| NC score | 0.027232 (rank : 92) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12860, Q12861, Q14030, Q8N466 | Gene names | CNTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3) (Glycoprotein gp135). | |||||
|
CNTN1_MOUSE
|
||||||
| NC score | 0.026748 (rank : 93) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12960, Q6NXV7, Q8BR42, Q8C6A0 | Gene names | Cntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3). | |||||
|
LSAMP_HUMAN
|
||||||
| NC score | 0.026653 (rank : 94) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13449, Q8IV49 | Gene names | LSAMP, LAMP | |||
|
Domain Architecture |

|
|||||
| Description | Limbic system-associated membrane protein precursor (LSAMP). | |||||
|
PHAR2_HUMAN
|
||||||
| NC score | 0.024110 (rank : 95) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
NGL1_HUMAN
|
||||||
| NC score | 0.023565 (rank : 96) | θ value | 0.00102713 (rank : 51) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCJ2, Q7L0N3 | Gene names | LRRC4C, KIAA1580, NGL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-G1 ligand precursor (NGL-1) (Leucine-rich repeat-containing protein 4C). | |||||
|
NGL1_MOUSE
|
||||||
| NC score | 0.023543 (rank : 97) | θ value | 0.000786445 (rank : 50) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 512 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8C031, Q8BGH8 | Gene names | Lrrc4c, Ngl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-G1 ligand precursor (NGL-1) (Leucine-rich repeat-containing protein 4C). | |||||
|
PXDC1_MOUSE
|
||||||
| NC score | 0.022709 (rank : 98) | θ value | 1.06291 (rank : 81) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91ZV7, Q8BM20, Q9CWV5 | Gene names | Plxdc1, Tem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7). | |||||
|
ESAM_MOUSE
|
||||||
| NC score | 0.022586 (rank : 99) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q925F2 | Gene names | Esam, Esam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.022205 (rank : 100) | θ value | 1.38821 (rank : 86) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.019579 (rank : 101) | θ value | 0.365318 (rank : 72) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.019465 (rank : 102) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
X3CL1_HUMAN
|
||||||
| NC score | 0.019207 (rank : 103) | θ value | 0.21417 (rank : 67) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78423, O00672 | Gene names | CX3CL1, A-152E5.2, FKN, NTT, SCYD1 | |||
|
Domain Architecture |

|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.019157 (rank : 104) | θ value | 0.163984 (rank : 64) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.017668 (rank : 105) | θ value | 0.62314 (rank : 77) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
RANB9_HUMAN
|
||||||
| NC score | 0.016448 (rank : 106) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
FMT_MOUSE
|
||||||
| NC score | 0.016207 (rank : 107) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D799, Q8VE89 | Gene names | Mtfmt, Fmt | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
LRC4B_MOUSE
|
||||||
| NC score | 0.016121 (rank : 108) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P0C192 | Gene names | Lrrc4b, Lrig4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 4B precursor. | |||||
|
LRC4B_HUMAN
|
||||||
| NC score | 0.015975 (rank : 109) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NT99, Q3ZCQ4, Q58F20 | Gene names | LRRC4B, LRIG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 4B precursor. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.015613 (rank : 110) | θ value | 0.365318 (rank : 70) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DRAP1_HUMAN
|
||||||
| NC score | 0.015388 (rank : 111) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14919, Q13448 | Gene names | DRAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.015274 (rank : 112) | θ value | 0.21417 (rank : 65) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.014610 (rank : 113) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
FMT_HUMAN
|
||||||
| NC score | 0.014127 (rank : 114) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DP5 | Gene names | MTFMT, FMT, FMT1 | |||
|
Domain Architecture |
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
CENG1_HUMAN
|
||||||
| NC score | 0.014090 (rank : 115) | θ value | 0.47712 (rank : 73) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
ATE1_MOUSE
|
||||||
| NC score | 0.012373 (rank : 116) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2A5, Q9Z2A4 | Gene names | Ate1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA--protein transferase 1 (EC 2.3.2.8) (R-transferase 1) (Arginyltransferase 1) (Arginine-tRNA--protein transferase 1). | |||||
|
YETS2_MOUSE
|
||||||
| NC score | 0.012323 (rank : 117) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
NEIL1_HUMAN
|
||||||
| NC score | 0.011745 (rank : 118) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96FI4, Q6ZRA7, Q86XW7, Q9H6C3 | Gene names | NEIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1) (FPG1). | |||||
|
IGSF2_HUMAN
|
||||||
| NC score | 0.011744 (rank : 119) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q93033, Q15856 | Gene names | IGSF2, CD101, EWI101, V7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin superfamily member 2 precursor (Glu-Trp-Ile EWI motif- containing protein 101) (EWI-101) (Cell surface glycoprotein V7) (CD101 antigen). | |||||
|
DRAP1_MOUSE
|
||||||
| NC score | 0.011536 (rank : 120) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D6N5 | Gene names | Drap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.011017 (rank : 121) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.009289 (rank : 122) | θ value | 1.06291 (rank : 80) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.009079 (rank : 123) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.008705 (rank : 124) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
PAPOA_HUMAN
|
||||||
| NC score | 0.007788 (rank : 125) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.007731 (rank : 126) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.007085 (rank : 127) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.007066 (rank : 128) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
SCG2_HUMAN
|
||||||
| NC score | 0.006621 (rank : 129) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.006251 (rank : 130) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
UTP20_MOUSE
|
||||||
| NC score | 0.005957 (rank : 131) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5XG71, Q80V22, Q8BXH9, Q8CHL6, Q99K11 | Gene names | Utp20, Drim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein). | |||||
|
CJ026_MOUSE
|
||||||
| NC score | 0.005841 (rank : 132) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGW2, Q3TF48, Q3U3M2, Q6PD43, Q8R0W8 | Gene names | D19Wsu162e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf26 homolog. | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.005825 (rank : 133) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.005759 (rank : 134) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
TEAD3_MOUSE
|
||||||
| NC score | 0.005604 (rank : 135) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70210, O08516, O70623, P70209 | Gene names | Tead3, Tcf13r2, Tef5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional enhancer factor TEF-5 (TEA domain family member 3) (TEAD-3) (ETF-related factor 1) (ETFR-1) (DTEF-1). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.004724 (rank : 136) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.003999 (rank : 137) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
VGFR2_HUMAN
|
||||||
| NC score | 0.003963 (rank : 138) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35968, O60723, Q14178 | Gene names | KDR, FLK1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 2 precursor (EC 2.7.10.1) (VEGFR-2) (Kinase insert domain receptor) (Protein-tyrosine kinase receptor Flk-1) (CD309 antigen). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.003850 (rank : 139) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.003806 (rank : 140) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HDAC6_HUMAN
|
||||||
| NC score | 0.003509 (rank : 141) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.003266 (rank : 142) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
TRPV1_MOUSE
|
||||||
| NC score | 0.003192 (rank : 143) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
FGFR3_MOUSE
|
||||||
| NC score | 0.003119 (rank : 144) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61851, Q61564, Q63834 | Gene names | Fgfr3, Mfr3, Sam3 | |||
|
Domain Architecture |

|
|||||
| Description | Fibroblast growth factor receptor 3 precursor (EC 2.7.10.1) (FGFR-3) (Heparin-binding growth factor receptor) (CD333 antigen). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.002535 (rank : 145) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ROR1_MOUSE
|
||||||
| NC score | 0.002519 (rank : 146) | θ value | 0.47712 (rank : 74) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z139 | Gene names | Ror1, Ntrkr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR1 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 1) (mROR1). | |||||
|
GP151_HUMAN
|
||||||
| NC score | 0.002475 (rank : 147) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDV0, Q86SN8, Q8NGV2 | Gene names | GPR151, PGR7 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 151 (G-protein coupled receptor PGR7) (GPCR-2037). | |||||
|
SNCAP_HUMAN
|
||||||
| NC score | 0.002417 (rank : 148) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
ZDHC8_MOUSE
|
||||||
| NC score | 0.002315 (rank : 149) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.001964 (rank : 150) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
DAF2_MOUSE
|
||||||
| NC score | 0.001647 (rank : 151) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61476 | Gene names | Cd55b, Daf2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor transmembrane isoform precursor (DAF-TM) (CD55 antigen). | |||||
|
TROP_HUMAN
|
||||||
| NC score | 0.001638 (rank : 152) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
SUHW4_HUMAN
|
||||||
| NC score | 0.000784 (rank : 153) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
ABI2_HUMAN
|
||||||
| NC score | 0.000498 (rank : 154) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
POGZ_MOUSE
|
||||||
| NC score | 0.000319 (rank : 155) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
POGZ_HUMAN
|
||||||
| NC score | -0.000114 (rank : 156) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z3K3, O75049, Q8TDZ7, Q9Y4X7 | Gene names | POGZ, KIAA0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
CHD4_MOUSE
|
||||||
| NC score | -0.000277 (rank : 157) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
GPR83_HUMAN
|
||||||
| NC score | -0.005654 (rank : 158) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 143 | Target Neighborhood Hits | 890 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYM4, Q9P1Y8 | Gene names | GPR83, GPR72, KIAA1540 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 83 precursor (G-protein coupled receptor 72). | |||||