Please be patient as the page loads
|
NEIL1_HUMAN
|
||||||
| SwissProt Accessions | Q96FI4, Q6ZRA7, Q86XW7, Q9H6C3 | Gene names | NEIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1) (FPG1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NEIL1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96FI4, Q6ZRA7, Q86XW7, Q9H6C3 | Gene names | NEIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1) (FPG1). | |||||
|
NEIL1_MOUSE
|
||||||
| θ value | 2.17438e-179 (rank : 2) | NC score | 0.971020 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K4Q6, Q80V58, Q9CYT9 | Gene names | Neil1, Nei1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1). | |||||
|
NEIL2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 3) | NC score | 0.296165 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969S2, Q7Z3Q7, Q8N842, Q8NG52 | Gene names | NEIL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
NEIL2_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 4) | NC score | 0.302674 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6R2P8 | Gene names | Neil2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
NEIL3_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 5) | NC score | 0.169489 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 6) | NC score | 0.056675 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.052555 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 8) | NC score | 0.058605 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.019792 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.018638 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.056429 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MYG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.052465 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JK81, Q9CYX0 | Gene names | Myg1 | |||
|
Domain Architecture |
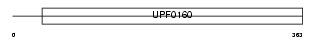
|
|||||
| Description | UPF0160 protein MYG1 (Protein Gamm1). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.042153 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.030035 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
IPR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.037852 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
PCAT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.031981 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NF37, Q1HAQ1, Q7Z4G6, Q8N3U7, Q8WUL8, Q9GZW6 | Gene names | AYTL2, LPCAT1, PFAAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (Acyltransferase-like 2) (Phosphonoformate immuno-associated protein 3). | |||||
|
PCAT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.031733 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TFD2, Q3TAX4, Q6NXZ6, Q8BG23, Q8BUX7, Q99JU6 | Gene names | Aytl2, Lpcat1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (mLPCAT1) (Acyltransferase-like 2). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.031743 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SEM4D_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.011745 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
USBP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.044712 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R370 | Gene names | Ushbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.016264 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
PRGR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.010581 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
BARX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.007327 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBU1 | Gene names | BARX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein BarH-like 1. | |||||
|
BASP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.027720 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.011281 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.015287 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.016008 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ZN192_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.000229 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15776, Q4VAR1, Q4VAR2, Q4VAR3, Q9H4T1 | Gene names | ZNF192 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 192 (LD5-1). | |||||
|
HCLS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.011824 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49710 | Gene names | Hcls1, Hs1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.015141 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.005175 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.021235 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.014882 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NUAK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | -0.000212 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||
|
TULP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.012621 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
NEIL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.079399 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96FI4, Q6ZRA7, Q86XW7, Q9H6C3 | Gene names | NEIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1) (FPG1). | |||||
|
NEIL1_MOUSE
|
||||||
| NC score | 0.971020 (rank : 2) | θ value | 2.17438e-179 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K4Q6, Q80V58, Q9CYT9 | Gene names | Neil1, Nei1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1). | |||||
|
NEIL2_MOUSE
|
||||||
| NC score | 0.302674 (rank : 3) | θ value | 0.00035302 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6R2P8 | Gene names | Neil2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
NEIL2_HUMAN
|
||||||
| NC score | 0.296165 (rank : 4) | θ value | 0.000270298 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969S2, Q7Z3Q7, Q8N842, Q8NG52 | Gene names | NEIL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
NEIL3_MOUSE
|
||||||
| NC score | 0.169489 (rank : 5) | θ value | 0.00175202 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL3_HUMAN
|
||||||
| NC score | 0.079399 (rank : 6) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.058605 (rank : 7) | θ value | 0.279714 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.056675 (rank : 8) | θ value | 0.0431538 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.056429 (rank : 9) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.052555 (rank : 10) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
MYG1_MOUSE
|
||||||
| NC score | 0.052465 (rank : 11) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JK81, Q9CYX0 | Gene names | Myg1 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0160 protein MYG1 (Protein Gamm1). | |||||
|
USBP1_MOUSE
|
||||||
| NC score | 0.044712 (rank : 12) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R370 | Gene names | Ushbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.042153 (rank : 13) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
IPR1_MOUSE
|
||||||
| NC score | 0.037852 (rank : 14) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
PCAT1_HUMAN
|
||||||
| NC score | 0.031981 (rank : 15) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NF37, Q1HAQ1, Q7Z4G6, Q8N3U7, Q8WUL8, Q9GZW6 | Gene names | AYTL2, LPCAT1, PFAAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (Acyltransferase-like 2) (Phosphonoformate immuno-associated protein 3). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.031743 (rank : 16) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PCAT1_MOUSE
|
||||||
| NC score | 0.031733 (rank : 17) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TFD2, Q3TAX4, Q6NXZ6, Q8BG23, Q8BUX7, Q99JU6 | Gene names | Aytl2, Lpcat1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (mLPCAT1) (Acyltransferase-like 2). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.030035 (rank : 18) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.027720 (rank : 19) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.021235 (rank : 20) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.019792 (rank : 21) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.018638 (rank : 22) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.016264 (rank : 23) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.016008 (rank : 24) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.015287 (rank : 25) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.015141 (rank : 26) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.014882 (rank : 27) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
TULP1_HUMAN
|
||||||
| NC score | 0.012621 (rank : 28) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
HCLS1_MOUSE
|
||||||
| NC score | 0.011824 (rank : 29) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49710 | Gene names | Hcls1, Hs1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1). | |||||
|
SEM4D_MOUSE
|
||||||
| NC score | 0.011745 (rank : 30) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.011281 (rank : 31) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
PRGR_HUMAN
|
||||||
| NC score | 0.010581 (rank : 32) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
BARX1_HUMAN
|
||||||
| NC score | 0.007327 (rank : 33) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBU1 | Gene names | BARX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein BarH-like 1. | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.005175 (rank : 34) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
ZN192_HUMAN
|
||||||
| NC score | 0.000229 (rank : 35) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15776, Q4VAR1, Q4VAR2, Q4VAR3, Q9H4T1 | Gene names | ZNF192 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 192 (LD5-1). | |||||
|
NUAK1_HUMAN
|
||||||
| NC score | -0.000212 (rank : 36) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||