Please be patient as the page loads
|
NEIL3_MOUSE
|
||||||
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NEIL3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.982026 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL2_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 3) | NC score | 0.469283 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q969S2, Q7Z3Q7, Q8N842, Q8NG52 | Gene names | NEIL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
TOP3A_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 4) | NC score | 0.336945 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13472, Q13473 | Gene names | TOP3A, TOP3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
TOP3A_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 5) | NC score | 0.338531 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70157 | Gene names | Top3a, Top3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
NEIL2_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 6) | NC score | 0.430401 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6R2P8 | Gene names | Neil2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
APEX2_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 7) | NC score | 0.341699 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68G58, Q8BJP7, Q8BTR7, Q8BUZ2, Q8BYE9, Q8R018, Q8R328, Q9CS12 | Gene names | Apex2, Ape2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2). | |||||
|
APEX2_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 8) | NC score | 0.329214 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBZ4, Q9Y5X7 | Gene names | APEX2, APE2, APEXL2, XTH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2) (APEX nuclease-like 2) (AP endonuclease XTH2). | |||||
|
NEIL1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 9) | NC score | 0.169489 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96FI4, Q6ZRA7, Q86XW7, Q9H6C3 | Gene names | NEIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1) (FPG1). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 10) | NC score | 0.109859 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
RBP2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 11) | NC score | 0.086484 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
RBP2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 12) | NC score | 0.090181 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
TTF2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 13) | NC score | 0.098608 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
ZRAN1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.150177 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
ZCHC4_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.141107 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BKW4, Q3UTX6, Q9D2I1 | Gene names | Zcchc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
NEIL1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.146027 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4Q6, Q80V58, Q9CYT9 | Gene names | Neil1, Nei1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1). | |||||
|
UB7I3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.092553 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYM8, O95623, Q96BS3, Q9BYM9 | Gene names | RBCK1, C20orf18, RNF54, UBCE7IP3, XAP4 | |||
|
Domain Architecture |
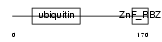
|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (Hepatitis B virus X- associated protein 4) (HBV-associated factor 4) (RING finger protein 54). | |||||
|
ZCHC4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.120720 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H5U6, Q5IW78, Q96AN7 | Gene names | ZCCHC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
TAB2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.062671 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYJ8, O94838, Q6I9W8, Q76N06, Q9UFP7 | Gene names | MAP3K7IP2, KIAA0733, TAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2). | |||||
|
UB7I3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.085922 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |
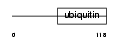
|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.017405 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.034492 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.020826 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.051399 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
GBRA4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.007351 (rank : 41) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D6F4 | Gene names | Gabra4 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit alpha-4 precursor (GABA(A) receptor subunit alpha-4). | |||||
|
MDM4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.041656 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35618 | Gene names | Mdm4, Mdmx | |||
|
Domain Architecture |
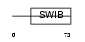
|
|||||
| Description | Mdm4 protein (p53-binding protein Mdm4) (Mdm2-like p53-binding protein) (Mdmx protein) (Double minute 4 protein). | |||||
|
STAB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.008581 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.014336 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
MDM4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.038966 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15151, Q6GS18, Q8IV83 | Gene names | MDM4, MDMX | |||
|
Domain Architecture |
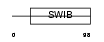
|
|||||
| Description | Mdm4 protein (p53-binding protein Mdm4) (Mdm2-like p53-binding protein) (Mdmx protein) (Double minute 4 protein). | |||||
|
ATS3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.005645 (rank : 43) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15072, Q9BXZ8 | Gene names | ADAMTS3, KIAA0366 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-3 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 3) (ADAM-TS 3) (ADAM-TS3) (Procollagen II amino propeptide-processing enzyme) (Procollagen II N-proteinase) (PC II-NP). | |||||
|
BIN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.029829 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQY0, Q9BVG2, Q9NVY9 | Gene names | BIN3 | |||
|
Domain Architecture |
|
|||||
| Description | Bridging integrator 3. | |||||
|
BIN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.027734 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI08 | Gene names | Bin3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bridging integrator 3. | |||||
|
NU153_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.076212 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
ITB8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.007765 (rank : 40) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
KR101_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.010037 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
RNF31_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.043724 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
BARD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.007042 (rank : 42) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
CCNL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.010508 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
KR109_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.009634 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
APEX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.056055 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27695, Q969L5, Q99775 | Gene names | APEX1, APE, APEX, APX, HAP1, REF1 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN) (REF-1 protein). | |||||
|
APEX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.055370 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28352 | Gene names | Apex1, Ape, Apex | |||
|
Domain Architecture |

|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN). | |||||
|
TOP3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.112883 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95985, Q9BUP5 | Gene names | TOP3B, TOP3B1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
TOP3B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.113250 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z321 | Gene names | Top3b, Top3b1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
NEIL3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL3_HUMAN
|
||||||
| NC score | 0.982026 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL2_HUMAN
|
||||||
| NC score | 0.469283 (rank : 3) | θ value | 7.09661e-13 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q969S2, Q7Z3Q7, Q8N842, Q8NG52 | Gene names | NEIL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
NEIL2_MOUSE
|
||||||
| NC score | 0.430401 (rank : 4) | θ value | 2.13673e-09 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6R2P8 | Gene names | Neil2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
APEX2_MOUSE
|
||||||
| NC score | 0.341699 (rank : 5) | θ value | 1.17247e-07 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68G58, Q8BJP7, Q8BTR7, Q8BUZ2, Q8BYE9, Q8R018, Q8R328, Q9CS12 | Gene names | Apex2, Ape2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2). | |||||
|
TOP3A_MOUSE
|
||||||
| NC score | 0.338531 (rank : 6) | θ value | 4.30538e-10 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70157 | Gene names | Top3a, Top3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
TOP3A_HUMAN
|
||||||
| NC score | 0.336945 (rank : 7) | θ value | 3.29651e-10 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13472, Q13473 | Gene names | TOP3A, TOP3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
APEX2_HUMAN
|
||||||
| NC score | 0.329214 (rank : 8) | θ value | 7.59969e-07 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBZ4, Q9Y5X7 | Gene names | APEX2, APE2, APEXL2, XTH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2) (APEX nuclease-like 2) (AP endonuclease XTH2). | |||||
|
NEIL1_HUMAN
|
||||||
| NC score | 0.169489 (rank : 9) | θ value | 0.00175202 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96FI4, Q6ZRA7, Q86XW7, Q9H6C3 | Gene names | NEIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1) (FPG1). | |||||
|
ZRAN1_HUMAN
|
||||||
| NC score | 0.150177 (rank : 10) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
NEIL1_MOUSE
|
||||||
| NC score | 0.146027 (rank : 11) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4Q6, Q80V58, Q9CYT9 | Gene names | Neil1, Nei1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1). | |||||
|
ZCHC4_MOUSE
|
||||||
| NC score | 0.141107 (rank : 12) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BKW4, Q3UTX6, Q9D2I1 | Gene names | Zcchc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
ZCHC4_HUMAN
|
||||||
| NC score | 0.120720 (rank : 13) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H5U6, Q5IW78, Q96AN7 | Gene names | ZCCHC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
TOP3B_MOUSE
|
||||||
| NC score | 0.113250 (rank : 14) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z321 | Gene names | Top3b, Top3b1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
TOP3B_HUMAN
|
||||||
| NC score | 0.112883 (rank : 15) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95985, Q9BUP5 | Gene names | TOP3B, TOP3B1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.109859 (rank : 16) | θ value | 0.00228821 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
TTF2_HUMAN
|
||||||
| NC score | 0.098608 (rank : 17) | θ value | 0.00869519 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
UB7I3_HUMAN
|
||||||
| NC score | 0.092553 (rank : 18) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYM8, O95623, Q96BS3, Q9BYM9 | Gene names | RBCK1, C20orf18, RNF54, UBCE7IP3, XAP4 | |||
|
Domain Architecture |

|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (Hepatitis B virus X- associated protein 4) (HBV-associated factor 4) (RING finger protein 54). | |||||
|
RBP2_HUMAN
|
||||||
| NC score | 0.090181 (rank : 19) | θ value | 0.00869519 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
RBP2_MOUSE
|
||||||
| NC score | 0.086484 (rank : 20) | θ value | 0.00390308 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
UB7I3_MOUSE
|
||||||
| NC score | 0.085922 (rank : 21) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |

|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.076212 (rank : 22) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
TAB2_HUMAN
|
||||||
| NC score | 0.062671 (rank : 23) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYJ8, O94838, Q6I9W8, Q76N06, Q9UFP7 | Gene names | MAP3K7IP2, KIAA0733, TAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2). | |||||
|
APEX1_HUMAN
|
||||||
| NC score | 0.056055 (rank : 24) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27695, Q969L5, Q99775 | Gene names | APEX1, APE, APEX, APX, HAP1, REF1 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN) (REF-1 protein). | |||||
|
APEX1_MOUSE
|
||||||
| NC score | 0.055370 (rank : 25) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28352 | Gene names | Apex1, Ape, Apex | |||
|
Domain Architecture |

|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN). | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.051399 (rank : 26) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
RNF31_HUMAN
|
||||||
| NC score | 0.043724 (rank : 27) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
MDM4_MOUSE
|
||||||
| NC score | 0.041656 (rank : 28) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35618 | Gene names | Mdm4, Mdmx | |||
|
Domain Architecture |

|
|||||
| Description | Mdm4 protein (p53-binding protein Mdm4) (Mdm2-like p53-binding protein) (Mdmx protein) (Double minute 4 protein). | |||||
|
MDM4_HUMAN
|
||||||
| NC score | 0.038966 (rank : 29) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15151, Q6GS18, Q8IV83 | Gene names | MDM4, MDMX | |||
|
Domain Architecture |

|
|||||
| Description | Mdm4 protein (p53-binding protein Mdm4) (Mdm2-like p53-binding protein) (Mdmx protein) (Double minute 4 protein). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.034492 (rank : 30) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
BIN3_HUMAN
|
||||||
| NC score | 0.029829 (rank : 31) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQY0, Q9BVG2, Q9NVY9 | Gene names | BIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Bridging integrator 3. | |||||
|
BIN3_MOUSE
|
||||||
| NC score | 0.027734 (rank : 32) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI08 | Gene names | Bin3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bridging integrator 3. | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.020826 (rank : 33) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.017405 (rank : 34) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.014336 (rank : 35) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
CCNL1_HUMAN
|
||||||
| NC score | 0.010508 (rank : 36) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.010037 (rank : 37) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.009634 (rank : 38) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.008581 (rank : 39) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
ITB8_HUMAN
|
||||||
| NC score | 0.007765 (rank : 40) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
GBRA4_MOUSE
|
||||||
| NC score | 0.007351 (rank : 41) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D6F4 | Gene names | Gabra4 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit alpha-4 precursor (GABA(A) receptor subunit alpha-4). | |||||
|
BARD1_HUMAN
|
||||||
| NC score | 0.007042 (rank : 42) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
ATS3_HUMAN
|
||||||
| NC score | 0.005645 (rank : 43) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15072, Q9BXZ8 | Gene names | ADAMTS3, KIAA0366 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-3 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 3) (ADAM-TS 3) (ADAM-TS3) (Procollagen II amino propeptide-processing enzyme) (Procollagen II N-proteinase) (PC II-NP). | |||||