Please be patient as the page loads
|
N4BP2_HUMAN
|
||||||
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
N4BP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
CN37_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 2) | NC score | 0.292222 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16330, Q61424, Q8C7C9, Q91V42, Q923F3 | Gene names | Cnp, Cnp1 | |||
|
Domain Architecture |
|
|||||
| Description | 2',3'-cyclic-nucleotide 3'-phosphodiesterase (EC 3.1.4.37) (CNP) (CNPase). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 3) | NC score | 0.176507 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
CN37_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 4) | NC score | 0.261259 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09543 | Gene names | CNP | |||
|
Domain Architecture |
|
|||||
| Description | 2',3'-cyclic-nucleotide 3'-phosphodiesterase (EC 3.1.4.37) (CNP) (CNPase). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 5) | NC score | 0.109785 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 6) | NC score | 0.144490 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.104008 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.076741 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.041332 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.092677 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.065724 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.090119 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
MGA_HUMAN
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.049414 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
ARHG1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.053191 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.084488 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TTF1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.078941 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15361, Q4VXF3, Q58EY2, Q6P5T5 | Gene names | TTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (RNA polymerase I termination factor). | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.063461 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.063436 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.067022 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
AIM1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.050717 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
DNAL4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.108126 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O96015 | Gene names | DNAL4 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein light chain 4, axonemal. | |||||
|
DNAL4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.108088 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCM4, O54793, Q3V233, Q99K65 | Gene names | Dnal4, Dnalc4 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein light chain 4, axonemal. | |||||
|
EXOC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.058756 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NV70, Q8WUE7, Q96T15, Q9NZE4 | Gene names | EXOC1, SEC3, SEC3L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
PCIF1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.063926 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59114 | Gene names | Pcif1 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.053855 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.039190 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
FXL13_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.041802 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEE6, Q6UVW7, Q6UVW8, Q75MN5, Q86UJ5, Q8N7Y4, Q8TCL2, Q8WUF9, Q8WUG0 | Gene names | FBXL13, FBL13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 13 (F-box and leucine-rich repeat protein 13). | |||||
|
DEK_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.069537 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.033380 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.060305 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.060314 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.053445 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.045503 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
ATL2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.020691 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TSK7, Q3U0C8 | Gene names | Adamtsl2, Kiaa0605, Tcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-like protein 2 precursor (ADAMTSL-2) (TSP1-repeats-containing protein 1) (TCP-1). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.038788 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.030695 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.061812 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.049184 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
NF2L2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.041537 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.022380 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.015366 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
DHB3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.031177 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70385 | Gene names | Hsd17b3, Edh17b3 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
LHX3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.019866 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
|
Domain Architecture |
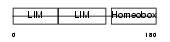
|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
LHX3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.019562 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50481, Q61800, Q61801 | Gene names | Lhx3, Lim-3, Lim3, Plim | |||
|
Domain Architecture |
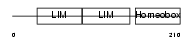
|
|||||
| Description | LIM/homeobox protein Lhx3 (Homeobox protein LIM-3) (Homeobox protein P-LIM) (LIM homeobox protein 3). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.034894 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.015335 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NRK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.054226 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWW6, Q8N430 | Gene names | NRK1, C9orf95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicotinamide riboside kinase 1 (EC 2.7.1.-). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.046063 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
E2F1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.028607 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01094, Q13143, Q92768 | Gene names | E2F1, RBBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F1 (E2F-1) (Retinoblastoma-binding protein 3) (RBBP-3) (PRB-binding protein E2F-1) (PBR3) (Retinoblastoma-associated protein 1) (RBAP-1). | |||||
|
MPP10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.043363 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.014587 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.023259 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.048949 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
DZIP3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.029163 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.082396 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.022451 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
ZN559_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.001019 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BR84 | Gene names | ZNF559 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 559. | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.022533 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CLIC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.023937 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1Q5 | Gene names | Clic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.051884 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.014020 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.057499 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.032810 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
TERA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.039164 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.039169 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.038375 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.043317 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NRK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.043802 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91W63, Q3TDW3 | Gene names | Nrk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicotinamide riboside kinase 1 (EC 2.7.1.-). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.030606 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
RGNEF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.033158 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
SPT6H_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.028385 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.013829 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.030395 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CF103_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.032833 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.037983 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.050245 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.021154 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.017959 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.011766 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TAIP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.019478 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59055 | Gene names | Taip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
4ET_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.028085 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
ARHGA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.021030 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
CLIC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.019539 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00299, Q15089 | Gene names | CLIC1, NCC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27) (Chloride channel ABP) (Regulatory nuclear chloride ion channel protein) (hRNCC). | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.040814 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
CP2E1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.004428 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05421, Q9Z198 | Gene names | Cyp2e1, Cyp2e, Cyp2e-1 | |||
|
Domain Architecture |
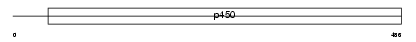
|
|||||
| Description | Cytochrome P450 2E1 (EC 1.14.14.1) (CYPIIE1) (P450-J) (P450-ALC). | |||||
|
KLC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.020571 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
KLC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.017772 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88447 | Gene names | Kns2, Klc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
MA2A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.014273 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16706, Q16767 | Gene names | MAN2A1, MANA2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2 (EC 3.2.1.114) (Alpha-mannosidase II) (Mannosyl- oligosaccharide 1,3-1,6-alpha-mannosidase) (MAN II) (Golgi alpha- mannosidase II) (Mannosidase alpha class 2A member 1). | |||||
|
PCX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.031380 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
SGOL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.026213 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.017533 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TOPRS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.022339 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
VDR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.006228 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48281 | Gene names | Vdr, Nr1i1 | |||
|
Domain Architecture |
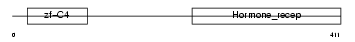
|
|||||
| Description | Vitamin D3 receptor (VDR) (1,25-dihydroxyvitamin D3 receptor). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.035836 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.023040 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CDR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.035715 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51861 | Gene names | CDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related antigen 1 (CDR34). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.020974 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
K0256_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.015724 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q93073, Q8N767 | Gene names | KIAA0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.027934 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
NELF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.019432 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99NF2, Q8BPT0, Q8C9R5, Q9DBF4, Q9ERZ1 | Gene names | Nelf, Jac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor) (Juxtasynaptic attractor of caldendrin on dendritic boutons protein) (Jacob protein). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.019802 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.032809 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.013238 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.035852 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
TLE6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.010265 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVB3 | Gene names | Tle6, Grg6 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 6 (Groucho-related protein 6). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.032066 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
ZN567_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | -0.000462 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N184, Q6N044 | Gene names | ZNF567 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 567. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.054246 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
N4BP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
CN37_MOUSE
|
||||||
| NC score | 0.292222 (rank : 2) | θ value | 3.41135e-07 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16330, Q61424, Q8C7C9, Q91V42, Q923F3 | Gene names | Cnp, Cnp1 | |||
|
Domain Architecture |

|
|||||
| Description | 2',3'-cyclic-nucleotide 3'-phosphodiesterase (EC 3.1.4.37) (CNP) (CNPase). | |||||
|
CN37_HUMAN
|
||||||
| NC score | 0.261259 (rank : 3) | θ value | 9.29e-05 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09543 | Gene names | CNP | |||
|
Domain Architecture |

|
|||||
| Description | 2',3'-cyclic-nucleotide 3'-phosphodiesterase (EC 3.1.4.37) (CNP) (CNPase). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.176507 (rank : 4) | θ value | 1.87187e-05 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.144490 (rank : 5) | θ value | 0.0113563 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.109785 (rank : 6) | θ value | 0.00298849 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DNAL4_HUMAN
|
||||||
| NC score | 0.108126 (rank : 7) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O96015 | Gene names | DNAL4 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein light chain 4, axonemal. | |||||
|
DNAL4_MOUSE
|
||||||
| NC score | 0.108088 (rank : 8) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCM4, O54793, Q3V233, Q99K65 | Gene names | Dnal4, Dnalc4 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein light chain 4, axonemal. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.104008 (rank : 9) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.092677 (rank : 10) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.090119 (rank : 11) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.084488 (rank : 12) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.082396 (rank : 13) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
TTF1_HUMAN
|
||||||
| NC score | 0.078941 (rank : 14) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15361, Q4VXF3, Q58EY2, Q6P5T5 | Gene names | TTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (RNA polymerase I termination factor). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.076741 (rank : 15) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.069537 (rank : 16) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.067022 (rank : 17) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.065724 (rank : 18) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
PCIF1_MOUSE
|
||||||
| NC score | 0.063926 (rank : 19) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59114 | Gene names | Pcif1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.063461 (rank : 20) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.063436 (rank : 21) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.061812 (rank : 22) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.060314 (rank : 23) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.060305 (rank : 24) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
EXOC1_HUMAN
|
||||||
| NC score | 0.058756 (rank : 25) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NV70, Q8WUE7, Q96T15, Q9NZE4 | Gene names | EXOC1, SEC3, SEC3L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.057499 (rank : 26) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.054246 (rank : 27) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
NRK1_HUMAN
|
||||||
| NC score | 0.054226 (rank : 28) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWW6, Q8N430 | Gene names | NRK1, C9orf95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicotinamide riboside kinase 1 (EC 2.7.1.-). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.053855 (rank : 29) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.053445 (rank : 30) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
ARHG1_HUMAN
|
||||||
| NC score | 0.053191 (rank : 31) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.051884 (rank : 32) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
AIM1_HUMAN
|
||||||
| NC score | 0.050717 (rank : 33) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.050245 (rank : 34) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
MGA_HUMAN
|
||||||
| NC score | 0.049414 (rank : 35) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.049184 (rank : 36) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.048949 (rank : 37) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.046063 (rank : 38) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.045503 (rank : 39) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
NRK1_MOUSE
|
||||||
| NC score | 0.043802 (rank : 40) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91W63, Q3TDW3 | Gene names | Nrk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicotinamide riboside kinase 1 (EC 2.7.1.-). | |||||
|
MPP10_MOUSE
|
||||||
| NC score | 0.043363 (rank : 41) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.043317 (rank : 42) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
FXL13_HUMAN
|
||||||
| NC score | 0.041802 (rank : 43) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEE6, Q6UVW7, Q6UVW8, Q75MN5, Q86UJ5, Q8N7Y4, Q8TCL2, Q8WUF9, Q8WUG0 | Gene names | FBXL13, FBL13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 13 (F-box and leucine-rich repeat protein 13). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.041537 (rank : 44) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.041332 (rank : 45) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.040814 (rank : 46) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.039190 (rank : 47) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
TERA_MOUSE
|
||||||
| NC score | 0.039169 (rank : 48) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_HUMAN
|
||||||
| NC score | 0.039164 (rank : 49) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.038788 (rank : 50) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.038375 (rank : 51) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.037983 (rank : 52) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.035852 (rank : 53) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.035836 (rank : 54) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
CDR1_HUMAN
|
||||||
| NC score | 0.035715 (rank : 55) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51861 | Gene names | CDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related antigen 1 (CDR34). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.034894 (rank : 56) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.033380 (rank : 57) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
RGNEF_MOUSE
|
||||||
| NC score | 0.033158 (rank : 58) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
CF103_HUMAN
|
||||||
| NC score | 0.032833 (rank : 59) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.032810 (rank : 60) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.032809 (rank : 61) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.032066 (rank : 62) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
PCX1_HUMAN
|
||||||
| NC score | 0.031380 (rank : 63) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
DHB3_MOUSE
|
||||||
| NC score | 0.031177 (rank : 64) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70385 | Gene names | Hsd17b3, Edh17b3 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.030695 (rank : 65) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.030606 (rank : 66) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.030395 (rank : 67) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
DZIP3_MOUSE
|
||||||
| NC score | 0.029163 (rank : 68) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
E2F1_HUMAN
|
||||||
| NC score | 0.028607 (rank : 69) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01094, Q13143, Q92768 | Gene names | E2F1, RBBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F1 (E2F-1) (Retinoblastoma-binding protein 3) (RBBP-3) (PRB-binding protein E2F-1) (PBR3) (Retinoblastoma-associated protein 1) (RBAP-1). | |||||
|
SPT6H_HUMAN
|
||||||
| NC score | 0.028385 (rank : 70) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
4ET_HUMAN
|
||||||
| NC score | 0.028085 (rank : 71) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.027934 (rank : 72) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
SGOL2_HUMAN
|
||||||
| NC score | 0.026213 (rank : 73) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
CLIC1_MOUSE
|
||||||
| NC score | 0.023937 (rank : 74) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1Q5 | Gene names | Clic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.023259 (rank : 75) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.023040 (rank : 76) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.022533 (rank : 77) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.022451 (rank : 78) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.022380 (rank : 79) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
TOPRS_HUMAN
|
||||||
| NC score | 0.022339 (rank : 80) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.021154 (rank : 81) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
ARHGA_HUMAN
|
||||||
| NC score | 0.021030 (rank : 82) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.020974 (rank : 83) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
ATL2_MOUSE
|
||||||
| NC score | 0.020691 (rank : 84) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TSK7, Q3U0C8 | Gene names | Adamtsl2, Kiaa0605, Tcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-like protein 2 precursor (ADAMTSL-2) (TSP1-repeats-containing protein 1) (TCP-1). | |||||
|
KLC1_HUMAN
|
||||||
| NC score | 0.020571 (rank : 85) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
LHX3_HUMAN
|
||||||
| NC score | 0.019866 (rank : 86) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.019802 (rank : 87) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
LHX3_MOUSE
|
||||||
| NC score | 0.019562 (rank : 88) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50481, Q61800, Q61801 | Gene names | Lhx3, Lim-3, Lim3, Plim | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx3 (Homeobox protein LIM-3) (Homeobox protein P-LIM) (LIM homeobox protein 3). | |||||
|
CLIC1_HUMAN
|
||||||
| NC score | 0.019539 (rank : 89) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00299, Q15089 | Gene names | CLIC1, NCC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27) (Chloride channel ABP) (Regulatory nuclear chloride ion channel protein) (hRNCC). | |||||
|
TAIP2_MOUSE
|
||||||
| NC score | 0.019478 (rank : 90) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59055 | Gene names | Taip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
NELF_MOUSE
|
||||||
| NC score | 0.019432 (rank : 91) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99NF2, Q8BPT0, Q8C9R5, Q9DBF4, Q9ERZ1 | Gene names | Nelf, Jac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor) (Juxtasynaptic attractor of caldendrin on dendritic boutons protein) (Jacob protein). | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.017959 (rank : 92) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
KLC1_MOUSE
|
||||||
| NC score | 0.017772 (rank : 93) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88447 | Gene names | Kns2, Klc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.017533 (rank : 94) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
K0256_HUMAN
|
||||||
| NC score | 0.015724 (rank : 95) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q93073, Q8N767 | Gene names | KIAA0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.015366 (rank : 96) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.015335 (rank : 97) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.014587 (rank : 98) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
MA2A1_HUMAN
|
||||||
| NC score | 0.014273 (rank : 99) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16706, Q16767 | Gene names | MAN2A1, MANA2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2 (EC 3.2.1.114) (Alpha-mannosidase II) (Mannosyl- oligosaccharide 1,3-1,6-alpha-mannosidase) (MAN II) (Golgi alpha- mannosidase II) (Mannosidase alpha class 2A member 1). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.014020 (rank : 100) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.013829 (rank : 101) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.013238 (rank : 102) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.011766 (rank : 103) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TLE6_MOUSE
|
||||||
| NC score | 0.010265 (rank : 104) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVB3 | Gene names | Tle6, Grg6 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 6 (Groucho-related protein 6). | |||||
|
VDR_MOUSE
|
||||||
| NC score | 0.006228 (rank : 105) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48281 | Gene names | Vdr, Nr1i1 | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin D3 receptor (VDR) (1,25-dihydroxyvitamin D3 receptor). | |||||
|
CP2E1_MOUSE
|
||||||
| NC score | 0.004428 (rank : 106) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05421, Q9Z198 | Gene names | Cyp2e1, Cyp2e, Cyp2e-1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2E1 (EC 1.14.14.1) (CYPIIE1) (P450-J) (P450-ALC). | |||||
|
ZN559_HUMAN
|
||||||
| NC score | 0.001019 (rank : 107) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BR84 | Gene names | ZNF559 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 559. | |||||
|
ZN567_HUMAN
|
||||||
| NC score | -0.000462 (rank : 108) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N184, Q6N044 | Gene names | ZNF567 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 567. | |||||