Please be patient as the page loads
|
RHBD4_MOUSE
|
||||||
| SwissProt Accessions | P58873 | Gene names | Rhbdl4, Vrho | |||
|
Domain Architecture |
|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RHBD4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998007 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P58872 | Gene names | RHBDL4, VRHO | |||
|
Domain Architecture |
|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
RHBD4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P58873 | Gene names | Rhbdl4, Vrho | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
RHBD1_MOUSE
|
||||||
| θ value | 1.17183e-124 (rank : 3) | NC score | 0.955613 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VC82 | Gene names | Rhbdl1, Rhbdl | |||
|
Domain Architecture |
|
|||||
| Description | Rhomboid-related protein 1 (EC 3.4.21.105) (RRP) (Rhomboid-like protein 1). | |||||
|
RHBD1_HUMAN
|
||||||
| θ value | 5.8157e-124 (rank : 4) | NC score | 0.951828 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75783, Q9NQ85 | Gene names | RHBDL1, RHBDL | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 1 (EC 3.4.21.105) (RRP) (Rhomboid-like protein 1). | |||||
|
RHBD2_HUMAN
|
||||||
| θ value | 5.38352e-45 (rank : 5) | NC score | 0.899101 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NX52, Q6P175, Q8NER8, Q96E02 | Gene names | RHBDL2 | |||
|
Domain Architecture |
|
|||||
| Description | Rhomboid-related protein 2 (EC 3.4.21.105) (RRP2) (Rhomboid-like protein 2). | |||||
|
CALM_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 6) | NC score | 0.178322 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 7) | NC score | 0.178322 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
GPDM_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 8) | NC score | 0.216101 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
PLSI_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 9) | NC score | 0.168758 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 10) | NC score | 0.175565 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
GPDM_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 11) | NC score | 0.213517 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
CABP7_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 12) | NC score | 0.194289 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |
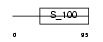
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 13) | NC score | 0.194289 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |
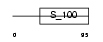
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
MLRA_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.151385 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |
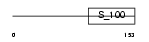
|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRA_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.150155 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q01449 | Gene names | MYL7, MYLC2A | |||
|
Domain Architecture |
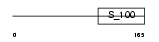
|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
CALM4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.168455 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
TNNC1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.163241 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |
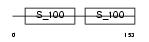
|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
EFHC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.102075 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5JVL4, Q5XKM4, Q6E1U7, Q6E1U8, Q8WUL2, Q9NVW6 | Gene names | EFHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing protein 1. | |||||
|
TNNC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.161956 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CABP2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.157489 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
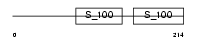
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
DJBP_MOUSE
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.124288 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
CABP2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.156325 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |
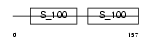
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
PLSL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.132679 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
PDCD6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.121812 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |
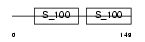
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.122015 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |
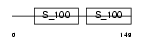
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.052322 (rank : 104) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.161215 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALL6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.168559 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
PLSL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.130339 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
PLST_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.131024 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
PLST_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.130664 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99K51 | Gene names | Pls3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
CAN9_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.037303 (rank : 109) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
DJBP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.125356 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
EFHC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.091691 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D9T8 | Gene names | Efhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing protein 1. | |||||
|
PRVA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.162285 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |
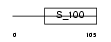
|
|||||
| Description | Parvalbumin alpha. | |||||
|
CABP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.157071 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |
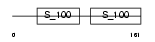
|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.156682 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |
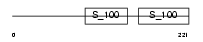
|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
ACTN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.064281 (rank : 91) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.152480 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
EFHA2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.048419 (rank : 108) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CTY5 | Gene names | Efha2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.049810 (rank : 107) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
CETN2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.143708 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |
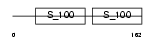
|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
EFCB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.136053 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
TNNC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.145921 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |
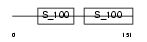
|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
ACTN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.061454 (rank : 92) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
CABP3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.146851 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |
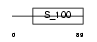
|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
EFCB3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.097160 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80X60 | Gene names | Efcab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 3. | |||||
|
TNNC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.145769 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |
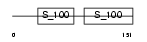
|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
AIF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.090301 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70200 | Gene names | Aif1, Iba1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Allograft inflammatory factor 1 (AIF-1) (Ionized calcium-binding adapter molecule 1). | |||||
|
CETN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.142203 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.006137 (rank : 114) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.026947 (rank : 111) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
ACTN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.056426 (rank : 100) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.056567 (rank : 99) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
EFCB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.088258 (rank : 66) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N7B9 | Gene names | EFCAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 3. | |||||
|
EFHA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.036890 (rank : 110) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86XE3, Q8IYZ3 | Gene names | EFHA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.013412 (rank : 112) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
CABP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.146867 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CETN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.139892 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
IBA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.072211 (rank : 87) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EQX4, Q8C150 | Gene names | Iba2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ionized calcium-binding adapter molecule 2. | |||||
|
S12A7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.008354 (rank : 113) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
AIF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.072145 (rank : 88) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P55008, Q9UKS9 | Gene names | AIF1, G1, IBA1 | |||
|
Domain Architecture |
|
|||||
| Description | Allograft inflammatory factor 1 (AIF-1) (Ionized calcium-binding adapter molecule 1) (Protein G1). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.132321 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.129647 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CALB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.058021 (rank : 96) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
CALB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.058531 (rank : 95) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.067774 (rank : 90) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.068096 (rank : 89) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CALN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.145680 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |
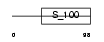
|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.145656 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |
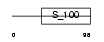
|
|||||
| Description | Calneuron-1. | |||||
|
CANB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.085929 (rank : 73) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.085937 (rank : 72) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |
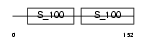
|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.080105 (rank : 82) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CANB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.088260 (rank : 65) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |
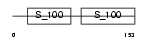
|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CAYP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.097900 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |
|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
CETN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.140104 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.130983 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.130872 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
CHP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.052250 (rank : 105) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P61022, Q62877, Q6ZWQ8 | Gene names | Chp, Sid470 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Sid 470). | |||||
|
CHP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.057997 (rank : 97) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
CHP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.056786 (rank : 98) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D869 | Gene names | Chp2, Hca520 | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520 homolog). | |||||
|
EFCB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.106747 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VUJ9, Q59G23, Q9BS36 | Gene names | EFCAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
EFHB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.060340 (rank : 93) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N7U6, Q6ZWK9, Q8IV58, Q96LQ6 | Gene names | EFHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
EFHD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.081826 (rank : 79) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BUP0, Q9BTF8, Q9H8I2, Q9HBQ0 | Gene names | EFHD1, SWS2 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
EFHD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.084391 (rank : 76) | |||
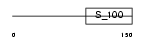
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D4J1, Q543U4 | Gene names | Efhd1, Sws2 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
EFHD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.079556 (rank : 83) | |||
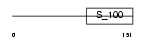
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96C19 | Gene names | EFHD2, SWS1 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
EFHD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.076095 (rank : 86) | |||
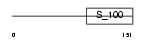
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D8Y0, Q9D0P4 | Gene names | Efhd2, Sws1 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
IBA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.058897 (rank : 94) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BQI0, Q6ZR40, Q8NAX7, Q8WU47, Q9H9G0 | Gene names | IBA2, C9orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ionized calcium-binding adapter molecule 2. | |||||
|
KIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.054789 (rank : 101) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75838 | Gene names | CIB2, KIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
KIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.054712 (rank : 102) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
KIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.050021 (rank : 106) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96Q77 | Gene names | CIB3, KIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 3 (Kinase-interacting protein 3) (KIP 3). | |||||
|
MLE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.084450 (rank : 75) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05976 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MLE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.081540 (rank : 81) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MLE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.083958 (rank : 77) | |||
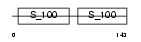
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P06741 | Gene names | MYL1 | |||
|
Domain Architecture |
|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MLE3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.081735 (rank : 80) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P05978 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |
|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MLR5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.105616 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |
|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
MLRM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.114313 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
MLRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.115116 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
MLRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.115386 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
MLRS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.086492 (rank : 71) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97457 | Gene names | Mylpf | |||
|
Domain Architecture |
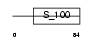
|
|||||
| Description | Myosin regulatory light chain 2, skeletal muscle isoform (MLC2F). | |||||
|
MLRV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.101163 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |
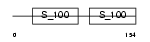
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLRV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.100222 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |
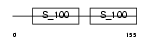
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MYL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.082328 (rank : 78) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08590, Q9NRS8 | Gene names | MYL3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain) (Cardiac myosin light chain-1) (CMLC1). | |||||
|
MYL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.084733 (rank : 74) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
MYL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.078202 (rank : 85) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12829, P11783 | Gene names | MYL4, MLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, embryonic muscle/atrial isoform) (Myosin light chain alkali, GT-1 isoform). | |||||
|
MYL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.079531 (rank : 84) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P09541 | Gene names | Myl4, Mlc1a, Myla | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, atrial/fetal isoform) (MLC1A) (MLC1EMB). | |||||
|
MYL6B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.089952 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
MYL6B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.087235 (rank : 69) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CI43 | Gene names | Myl6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B). | |||||
|
MYL6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.087755 (rank : 67) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |
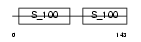
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.087755 (rank : 68) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
ONCO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.107235 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P32930 | Gene names | OCM, OCMN | |||
|
Domain Architecture |
|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
ONCO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.087089 (rank : 70) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P51879, Q62004 | Gene names | Ocm | |||
|
Domain Architecture |
|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
PRVA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.116594 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20472, P78378, Q4VB78, Q5R3Q9 | Gene names | PVALB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parvalbumin alpha. | |||||
|
SEGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.052772 (rank : 103) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |
|
|||||
| Description | Secretagogin. | |||||
|
RHBD4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P58873 | Gene names | Rhbdl4, Vrho | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
RHBD4_HUMAN
|
||||||
| NC score | 0.998007 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P58872 | Gene names | RHBDL4, VRHO | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
RHBD1_MOUSE
|
||||||
| NC score | 0.955613 (rank : 3) | θ value | 1.17183e-124 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VC82 | Gene names | Rhbdl1, Rhbdl | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 1 (EC 3.4.21.105) (RRP) (Rhomboid-like protein 1). | |||||
|
RHBD1_HUMAN
|
||||||
| NC score | 0.951828 (rank : 4) | θ value | 5.8157e-124 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75783, Q9NQ85 | Gene names | RHBDL1, RHBDL | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 1 (EC 3.4.21.105) (RRP) (Rhomboid-like protein 1). | |||||
|
RHBD2_HUMAN
|
||||||
| NC score | 0.899101 (rank : 5) | θ value | 5.38352e-45 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NX52, Q6P175, Q8NER8, Q96E02 | Gene names | RHBDL2 | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 2 (EC 3.4.21.105) (RRP2) (Rhomboid-like protein 2). | |||||
|
GPDM_MOUSE
|
||||||
| NC score | 0.216101 (rank : 6) | θ value | 0.000461057 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
GPDM_HUMAN
|
||||||
| NC score | 0.213517 (rank : 7) | θ value | 0.000786445 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
CABP7_HUMAN
|
||||||
| NC score | 0.194289 (rank : 8) | θ value | 0.0113563 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| NC score | 0.194289 (rank : 9) | θ value | 0.0113563 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CALM_HUMAN
|
||||||
| NC score | 0.178322 (rank : 10) | θ value | 0.00020696 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| NC score | 0.178322 (rank : 11) | θ value | 0.00020696 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.175565 (rank : 12) | θ value | 0.000786445 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
PLSI_HUMAN
|
||||||
| NC score | 0.168758 (rank : 13) | θ value | 0.000602161 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
CALL6_HUMAN
|
||||||
| NC score | 0.168559 (rank : 14) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.168455 (rank : 15) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
TNNC1_MOUSE
|
||||||
| NC score | 0.163241 (rank : 16) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
PRVA_MOUSE
|
||||||
| NC score | 0.162285 (rank : 17) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |

|
|||||
| Description | Parvalbumin alpha. | |||||
|
TNNC1_HUMAN
|
||||||
| NC score | 0.161956 (rank : 18) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.161215 (rank : 19) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CABP2_HUMAN
|
||||||
| NC score | 0.157489 (rank : 20) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
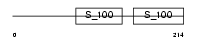
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP1_HUMAN
|
||||||
| NC score | 0.157071 (rank : 21) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP1_MOUSE
|
||||||
| NC score | 0.156682 (rank : 22) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CABP2_MOUSE
|
||||||
| NC score | 0.156325 (rank : 23) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.152480 (rank : 24) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
MLRA_MOUSE
|
||||||
| NC score | 0.151385 (rank : 25) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRA_HUMAN
|
||||||
| NC score | 0.150155 (rank : 26) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q01449 | Gene names | MYL7, MYLC2A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.146867 (rank : 27) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP3_HUMAN
|
||||||
| NC score | 0.146851 (rank : 28) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
TNNC2_MOUSE
|
||||||
| NC score | 0.145921 (rank : 29) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
TNNC2_HUMAN
|
||||||
| NC score | 0.145769 (rank : 30) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
CALN1_HUMAN
|
||||||
| NC score | 0.145680 (rank : 31) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| NC score | 0.145656 (rank : 32) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1. | |||||
|
CETN2_HUMAN
|
||||||
| NC score | 0.143708 (rank : 33) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN2_MOUSE
|
||||||
| NC score | 0.142203 (rank : 34) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN1_HUMAN
|
||||||
| NC score | 0.140104 (rank : 35) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN1_MOUSE
|
||||||
| NC score | 0.139892 (rank : 36) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
EFCB2_MOUSE
|
||||||
| NC score | 0.136053 (rank : 37) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
PLSL_MOUSE
|
||||||
| NC score | 0.132679 (rank : 38) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.132321 (rank : 39) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
PLST_HUMAN
|
||||||
| NC score | 0.131024 (rank : 40) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
CETN3_HUMAN
|
||||||
| NC score | 0.130983 (rank : 41) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| NC score | 0.130872 (rank : 42) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
PLST_MOUSE
|
||||||
| NC score | 0.130664 (rank : 43) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99K51 | Gene names | Pls3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
PLSL_HUMAN
|
||||||
| NC score | 0.130339 (rank : 44) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
CABP4_MOUSE
|
||||||
| NC score | 0.129647 (rank : 45) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
DJBP_HUMAN
|
||||||
| NC score | 0.125356 (rank : 46) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
DJBP_MOUSE
|
||||||
| NC score | 0.124288 (rank : 47) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
PDCD6_MOUSE
|
||||||
| NC score | 0.122015 (rank : 48) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
PDCD6_HUMAN
|
||||||
| NC score | 0.121812 (rank : 49) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PRVA_HUMAN
|
||||||
| NC score | 0.116594 (rank : 50) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20472, P78378, Q4VB78, Q5R3Q9 | Gene names | PVALB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parvalbumin alpha. | |||||
|
MLRN_MOUSE
|
||||||
| NC score | 0.115386 (rank : 51) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
MLRN_HUMAN
|
||||||
| NC score | 0.115116 (rank : 52) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
MLRM_HUMAN
|
||||||
| NC score | 0.114313 (rank : 53) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
ONCO_HUMAN
|
||||||
| NC score | 0.107235 (rank : 54) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P32930 | Gene names | OCM, OCMN | |||
|
Domain Architecture |

|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
EFCB2_HUMAN
|
||||||
| NC score | 0.106747 (rank : 55) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VUJ9, Q59G23, Q9BS36 | Gene names | EFCAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
MLR5_HUMAN
|
||||||
| NC score | 0.105616 (rank : 56) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |

|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
EFHC1_HUMAN
|
||||||
| NC score | 0.102075 (rank : 57) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5JVL4, Q5XKM4, Q6E1U7, Q6E1U8, Q8WUL2, Q9NVW6 | Gene names | EFHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing protein 1. | |||||
|
MLRV_HUMAN
|
||||||
| NC score | 0.101163 (rank : 58) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLRV_MOUSE
|
||||||
| NC score | 0.100222 (rank : 59) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
CAYP1_HUMAN
|
||||||
| NC score | 0.097900 (rank : 60) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
EFCB3_MOUSE
|
||||||
| NC score | 0.097160 (rank : 61) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80X60 | Gene names | Efcab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 3. | |||||
|
EFHC1_MOUSE
|
||||||
| NC score | 0.091691 (rank : 62) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D9T8 | Gene names | Efhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing protein 1. | |||||
|
AIF1_MOUSE
|
||||||
| NC score | 0.090301 (rank : 63) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70200 | Gene names | Aif1, Iba1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Allograft inflammatory factor 1 (AIF-1) (Ionized calcium-binding adapter molecule 1). | |||||
|
MYL6B_HUMAN
|
||||||
| NC score | 0.089952 (rank : 64) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
CANB2_MOUSE
|
||||||
| NC score | 0.088260 (rank : 65) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
EFCB3_HUMAN
|
||||||
| NC score | 0.088258 (rank : 66) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N7B9 | Gene names | EFCAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 3. | |||||
|
MYL6_HUMAN
|
||||||
| NC score | 0.087755 (rank : 67) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| NC score | 0.087755 (rank : 68) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6B_MOUSE
|
||||||
| NC score | 0.087235 (rank : 69) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CI43 | Gene names | Myl6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B). | |||||
|
ONCO_MOUSE
|
||||||
| NC score | 0.087089 (rank : 70) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P51879, Q62004 | Gene names | Ocm | |||
|
Domain Architecture |

|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
MLRS_MOUSE
|
||||||
| NC score | 0.086492 (rank : 71) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97457 | Gene names | Mylpf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, skeletal muscle isoform (MLC2F). | |||||
|
CANB1_MOUSE
|
||||||
| NC score | 0.085937 (rank : 72) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_HUMAN
|
||||||
| NC score | 0.085929 (rank : 73) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
MYL3_MOUSE
|
||||||
| NC score | 0.084733 (rank : 74) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
MLE1_HUMAN
|
||||||
| NC score | 0.084450 (rank : 75) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05976 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
EFHD1_MOUSE
|
||||||
| NC score | 0.084391 (rank : 76) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D4J1, Q543U4 | Gene names | Efhd1, Sws2 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
MLE3_HUMAN
|
||||||
| NC score | 0.083958 (rank : 77) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P06741 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MYL3_HUMAN
|
||||||
| NC score | 0.082328 (rank : 78) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08590, Q9NRS8 | Gene names | MYL3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain) (Cardiac myosin light chain-1) (CMLC1). | |||||
|
EFHD1_HUMAN
|
||||||
| NC score | 0.081826 (rank : 79) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BUP0, Q9BTF8, Q9H8I2, Q9HBQ0 | Gene names | EFHD1, SWS2 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
MLE3_MOUSE
|
||||||
| NC score | 0.081735 (rank : 80) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P05978 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MLE1_MOUSE
|
||||||
| NC score | 0.081540 (rank : 81) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
CANB2_HUMAN
|
||||||
| NC score | 0.080105 (rank : 82) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
EFHD2_HUMAN
|
||||||
| NC score | 0.079556 (rank : 83) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96C19 | Gene names | EFHD2, SWS1 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
MYL4_MOUSE
|
||||||
| NC score | 0.079531 (rank : 84) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P09541 | Gene names | Myl4, Mlc1a, Myla | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, atrial/fetal isoform) (MLC1A) (MLC1EMB). | |||||
|
MYL4_HUMAN
|
||||||
| NC score | 0.078202 (rank : 85) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12829, P11783 | Gene names | MYL4, MLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, embryonic muscle/atrial isoform) (Myosin light chain alkali, GT-1 isoform). | |||||
|
EFHD2_MOUSE
|
||||||
| NC score | 0.076095 (rank : 86) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D8Y0, Q9D0P4 | Gene names | Efhd2, Sws1 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
IBA2_MOUSE
|
||||||
| NC score | 0.072211 (rank : 87) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EQX4, Q8C150 | Gene names | Iba2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ionized calcium-binding adapter molecule 2. | |||||
|
AIF1_HUMAN
|
||||||
| NC score | 0.072145 (rank : 88) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P55008, Q9UKS9 | Gene names | AIF1, G1, IBA1 | |||
|
Domain Architecture |

|
|||||
| Description | Allograft inflammatory factor 1 (AIF-1) (Ionized calcium-binding adapter molecule 1) (Protein G1). | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.068096 (rank : 89) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.067774 (rank : 90) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
ACTN2_HUMAN
|
||||||
| NC score | 0.064281 (rank : 91) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| NC score | 0.061454 (rank : 92) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
EFHB_HUMAN
|
||||||
| NC score | 0.060340 (rank : 93) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N7U6, Q6ZWK9, Q8IV58, Q96LQ6 | Gene names | EFHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
IBA2_HUMAN
|
||||||
| NC score | 0.058897 (rank : 94) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BQI0, Q6ZR40, Q8NAX7, Q8WU47, Q9H9G0 | Gene names | IBA2, C9orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ionized calcium-binding adapter molecule 2. | |||||
|
CALB1_MOUSE
|
||||||
| NC score | 0.058531 (rank : 95) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
CALB1_HUMAN
|
||||||
| NC score | 0.058021 (rank : 96) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
CHP2_HUMAN
|
||||||
| NC score | 0.057997 (rank : 97) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
CHP2_MOUSE
|
||||||
| NC score | 0.056786 (rank : 98) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D869 | Gene names | Chp2, Hca520 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520 homolog). | |||||
|
ACTN3_MOUSE
|
||||||
| NC score | 0.056567 (rank : 99) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_HUMAN
|
||||||
| NC score | 0.056426 (rank : 100) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
KIP2_HUMAN
|
||||||
| NC score | 0.054789 (rank : 101) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75838 | Gene names | CIB2, KIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
KIP2_MOUSE
|
||||||
| NC score | 0.054712 (rank : 102) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
SEGN_HUMAN
|
||||||
| NC score | 0.052772 (rank : 103) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.052322 (rank : 104) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
CHP1_MOUSE
|
||||||
| NC score | 0.052250 (rank : 105) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P61022, Q62877, Q6ZWQ8 | Gene names | Chp, Sid470 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Sid 470). | |||||
|
KIP3_HUMAN
|
||||||
| NC score | 0.050021 (rank : 106) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96Q77 | Gene names | CIB3, KIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 3 (Kinase-interacting protein 3) (KIP 3). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.049810 (rank : 107) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
EFHA2_MOUSE
|
||||||
| NC score | 0.048419 (rank : 108) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CTY5 | Gene names | Efha2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
CAN9_MOUSE
|
||||||
| NC score | 0.037303 (rank : 109) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
EFHA2_HUMAN
|
||||||
| NC score | 0.036890 (rank : 110) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86XE3, Q8IYZ3 | Gene names | EFHA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.026947 (rank : 111) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.013412 (rank : 112) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
S12A7_MOUSE
|
||||||
| NC score | 0.008354 (rank : 113) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.006137 (rank : 114) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||