Please be patient as the page loads
|
ATG9A_HUMAN
|
||||||
| SwissProt Accessions | Q7Z3C6, Q6P0N7, Q7Z317, Q7Z320, Q8NDK6, Q8WU65, Q9BVL5, Q9H6L1, Q9HAG7 | Gene names | ATG9A, APG9L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 9A (APG9-like 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ATG9A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z3C6, Q6P0N7, Q7Z317, Q7Z320, Q8NDK6, Q8WU65, Q9BVL5, Q9H6L1, Q9HAG7 | Gene names | ATG9A, APG9L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 9A (APG9-like 1). | |||||
|
ATG9A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.974274 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FE2 | Gene names | Atg9a, Apg9l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 9A (APG9-like 1). | |||||
|
CK056_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 3) | NC score | 0.107180 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U2I3, Q3TWF5, Q6P543, Q80T93, Q811J6, Q8BI34, Q8VE55 | Gene names | Kiaa1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56 homolog. | |||||
|
DYN1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 4) | NC score | 0.041758 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
IKBE_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.030651 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00221 | Gene names | NFKBIE, IKBE | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKBE). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 0.125558 (rank : 6) | NC score | 0.017576 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 7) | NC score | 0.056377 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.039339 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.032777 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
PDPK1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.005228 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2A0, Q9R1D8, Q9R215 | Gene names | Pdpk1, Pdk1 | |||
|
Domain Architecture |
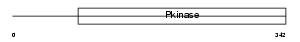
|
|||||
| Description | 3-phosphoinositide-dependent protein kinase 1 (EC 2.7.11.1) (mPDK1). | |||||
|
CK056_HUMAN
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.083653 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N612, Q9C0A4, Q9H0N3, Q9H624 | Gene names | C11orf56, KIAA1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56. | |||||
|
EGLN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.030168 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
KIF1C_MOUSE
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.012676 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
PRDM8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.029664 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NQV8, Q6IQ36 | Gene names | PRDM8, PFM5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 8 (PR domain-containing protein 8). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.036148 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
CLAP2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.030028 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.015516 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.036853 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
KLF14_HUMAN
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.007701 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TD94 | Gene names | KLF14, BTEB5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 14 (Transcription factor BTEB5) (Basic transcription element-binding protein 5) (BTE-binding protein 5). | |||||
|
PAR6G_MOUSE
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.024456 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JK84 | Gene names | Pard6g, Par6g | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog gamma (PAR-6 gamma) (PAR6A). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.039870 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.023427 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.025903 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
MGA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.021257 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.014972 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
PUM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.028613 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80U58, Q80UZ9, Q91YW4, Q925A0, Q9ERC7 | Gene names | Pum2, Kiaa0235 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 2. | |||||
|
TF2AA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.041347 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PM3, Q8C812 | Gene names | Gtf2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
PAXI_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.011650 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49023, O14970, O14971, O60360 | Gene names | PXN | |||
|
Domain Architecture |

|
|||||
| Description | Paxillin. | |||||
|
PRD16_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.005420 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HAZ2, Q8WYJ9, Q9C0I8 | Gene names | PRDM16, KIAA1675, MEL1, PFM13 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 16 (PR domain-containing protein 16) (Transcription factor MEL1). | |||||
|
TM16H_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.025584 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.029736 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DVL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.014279 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.020053 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.023388 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ZBT46_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.006696 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BID6, Q3U2R2, Q9D0G6, Q9D492 | Gene names | Zbtb46, Btbd4 | |||
|
Domain Architecture |
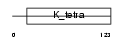
|
|||||
| Description | Zinc finger and BTB domain-containing protein 46 (BTB/POZ domain- containing protein 4). | |||||
|
ZIC5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.005400 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96T25 | Gene names | ZIC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 5 (Zinc finger protein of the cerebellum 5). | |||||
|
BAG4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.022170 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CI61, Q3TRL9, Q91VT5, Q9CWG2 | Gene names | Bag4, Sodd | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.013278 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.012845 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.011296 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
DNER_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.004205 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
GLI4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.002177 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 735 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10075, Q96CK9 | Gene names | GLI4, HKR4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein GLI4 (Krueppel-related zinc finger protein 4) (Protein HKR4). | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.032721 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.018059 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.016589 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
IRX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.010419 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.022859 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.016670 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SKIV2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.016910 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15477, O15005, Q12902, Q15476 | Gene names | SKIV2L, DDX13, SKI2W, SKIV2, W | |||
|
Domain Architecture |

|
|||||
| Description | Helicase SKI2W (EC 3.6.1.-) (Helicase-like protein) (HLP). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.018012 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TF2AA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.032829 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52655 | Gene names | GTF2A1, TF2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) (TFIIA-42) (TFIIAL) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
CO1A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.009518 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
H6ST3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.011452 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.023918 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
OLIG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.009940 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
PUM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.023156 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TB72, O00234, Q53TV7, Q8WY43, Q9HAN2 | Gene names | PUM2, KIAA0235, PUMH2 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 2 (Pumilio-2). | |||||
|
VASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.012888 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
WDR72_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.016788 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3MJ13, Q7Z3I3, Q8N8X2 | Gene names | WDR72 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 72. | |||||
|
ATG9A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z3C6, Q6P0N7, Q7Z317, Q7Z320, Q8NDK6, Q8WU65, Q9BVL5, Q9H6L1, Q9HAG7 | Gene names | ATG9A, APG9L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 9A (APG9-like 1). | |||||
|
ATG9A_MOUSE
|
||||||
| NC score | 0.974274 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FE2 | Gene names | Atg9a, Apg9l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 9A (APG9-like 1). | |||||
|
CK056_MOUSE
|
||||||
| NC score | 0.107180 (rank : 3) | θ value | 0.0148317 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U2I3, Q3TWF5, Q6P543, Q80T93, Q811J6, Q8BI34, Q8VE55 | Gene names | Kiaa1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56 homolog. | |||||
|
CK056_HUMAN
|
||||||
| NC score | 0.083653 (rank : 4) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N612, Q9C0A4, Q9H0N3, Q9H624 | Gene names | C11orf56, KIAA1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.056377 (rank : 5) | θ value | 0.125558 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
DYN1_HUMAN
|
||||||
| NC score | 0.041758 (rank : 6) | θ value | 0.0431538 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
TF2AA_MOUSE
|
||||||
| NC score | 0.041347 (rank : 7) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PM3, Q8C812 | Gene names | Gtf2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.039870 (rank : 8) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.039339 (rank : 9) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.036853 (rank : 10) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.036148 (rank : 11) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
TF2AA_HUMAN
|
||||||
| NC score | 0.032829 (rank : 12) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52655 | Gene names | GTF2A1, TF2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) (TFIIA-42) (TFIIAL) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.032777 (rank : 13) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.032721 (rank : 14) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
IKBE_HUMAN
|
||||||
| NC score | 0.030651 (rank : 15) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00221 | Gene names | NFKBIE, IKBE | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKBE). | |||||
|
EGLN1_MOUSE
|
||||||
| NC score | 0.030168 (rank : 16) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
CLAP2_MOUSE
|
||||||
| NC score | 0.030028 (rank : 17) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.029736 (rank : 18) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PRDM8_HUMAN
|
||||||
| NC score | 0.029664 (rank : 19) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NQV8, Q6IQ36 | Gene names | PRDM8, PFM5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 8 (PR domain-containing protein 8). | |||||
|
PUM2_MOUSE
|
||||||
| NC score | 0.028613 (rank : 20) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80U58, Q80UZ9, Q91YW4, Q925A0, Q9ERC7 | Gene names | Pum2, Kiaa0235 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 2. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.025903 (rank : 21) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TM16H_HUMAN
|
||||||
| NC score | 0.025584 (rank : 22) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
PAR6G_MOUSE
|
||||||
| NC score | 0.024456 (rank : 23) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JK84 | Gene names | Pard6g, Par6g | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog gamma (PAR-6 gamma) (PAR6A). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.023918 (rank : 24) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.023427 (rank : 25) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.023388 (rank : 26) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
PUM2_HUMAN
|
||||||
| NC score | 0.023156 (rank : 27) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TB72, O00234, Q53TV7, Q8WY43, Q9HAN2 | Gene names | PUM2, KIAA0235, PUMH2 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 2 (Pumilio-2). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.022859 (rank : 28) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
BAG4_MOUSE
|
||||||
| NC score | 0.022170 (rank : 29) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CI61, Q3TRL9, Q91VT5, Q9CWG2 | Gene names | Bag4, Sodd | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
MGA_HUMAN
|
||||||
| NC score | 0.021257 (rank : 30) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.020053 (rank : 31) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.018059 (rank : 32) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.018012 (rank : 33) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.017576 (rank : 34) | θ value | 0.125558 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
SKIV2_HUMAN
|
||||||
| NC score | 0.016910 (rank : 35) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15477, O15005, Q12902, Q15476 | Gene names | SKIV2L, DDX13, SKI2W, SKIV2, W | |||
|
Domain Architecture |

|
|||||
| Description | Helicase SKI2W (EC 3.6.1.-) (Helicase-like protein) (HLP). | |||||
|
WDR72_HUMAN
|
||||||
| NC score | 0.016788 (rank : 36) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3MJ13, Q7Z3I3, Q8N8X2 | Gene names | WDR72 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 72. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.016670 (rank : 37) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.016589 (rank : 38) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.015516 (rank : 39) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.014972 (rank : 40) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
DVL2_MOUSE
|
||||||
| NC score | 0.014279 (rank : 41) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.013278 (rank : 42) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.012888 (rank : 43) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.012845 (rank : 44) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
KIF1C_MOUSE
|
||||||
| NC score | 0.012676 (rank : 45) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
PAXI_HUMAN
|
||||||
| NC score | 0.011650 (rank : 46) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49023, O14970, O14971, O60360 | Gene names | PXN | |||
|
Domain Architecture |

|
|||||
| Description | Paxillin. | |||||
|
H6ST3_HUMAN
|
||||||
| NC score | 0.011452 (rank : 47) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.011296 (rank : 48) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
IRX2_HUMAN
|
||||||
| NC score | 0.010419 (rank : 49) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
OLIG2_HUMAN
|
||||||
| NC score | 0.009940 (rank : 50) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
CO1A1_MOUSE
|
||||||
| NC score | 0.009518 (rank : 51) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
KLF14_HUMAN
|
||||||
| NC score | 0.007701 (rank : 52) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TD94 | Gene names | KLF14, BTEB5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 14 (Transcription factor BTEB5) (Basic transcription element-binding protein 5) (BTE-binding protein 5). | |||||
|
ZBT46_MOUSE
|
||||||
| NC score | 0.006696 (rank : 53) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BID6, Q3U2R2, Q9D0G6, Q9D492 | Gene names | Zbtb46, Btbd4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 46 (BTB/POZ domain- containing protein 4). | |||||
|
PRD16_HUMAN
|
||||||
| NC score | 0.005420 (rank : 54) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HAZ2, Q8WYJ9, Q9C0I8 | Gene names | PRDM16, KIAA1675, MEL1, PFM13 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 16 (PR domain-containing protein 16) (Transcription factor MEL1). | |||||
|
ZIC5_HUMAN
|
||||||
| NC score | 0.005400 (rank : 55) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96T25 | Gene names | ZIC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 5 (Zinc finger protein of the cerebellum 5). | |||||
|
PDPK1_MOUSE
|
||||||
| NC score | 0.005228 (rank : 56) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2A0, Q9R1D8, Q9R215 | Gene names | Pdpk1, Pdk1 | |||
|
Domain Architecture |

|
|||||
| Description | 3-phosphoinositide-dependent protein kinase 1 (EC 2.7.11.1) (mPDK1). | |||||
|
DNER_HUMAN
|
||||||
| NC score | 0.004205 (rank : 57) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
GLI4_HUMAN
|
||||||
| NC score | 0.002177 (rank : 58) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 735 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10075, Q96CK9 | Gene names | GLI4, HKR4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein GLI4 (Krueppel-related zinc finger protein 4) (Protein HKR4). | |||||