Please be patient as the page loads
|
NKRF_MOUSE
|
||||||
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NKRF_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994796 (rank : 2) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
NKRF_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
GPTC2_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 3) | NC score | 0.397540 (rank : 3) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 4) | NC score | 0.375008 (rank : 6) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SFR14_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 5) | NC score | 0.386732 (rank : 5) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
GPTC2_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 6) | NC score | 0.388514 (rank : 4) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
AGGF1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 7) | NC score | 0.361850 (rank : 7) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
SF04_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 8) | NC score | 0.349765 (rank : 8) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SF04_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 9) | NC score | 0.342777 (rank : 9) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 10) | NC score | 0.340282 (rank : 10) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
RBM5_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 11) | NC score | 0.298629 (rank : 11) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 12) | NC score | 0.298004 (rank : 12) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
RBM10_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 13) | NC score | 0.292343 (rank : 13) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
SMBP2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 14) | NC score | 0.118952 (rank : 37) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 15) | NC score | 0.106619 (rank : 38) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 16) | NC score | 0.152779 (rank : 33) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
VPK1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.233680 (rank : 19) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK7_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 18) | NC score | 0.229672 (rank : 21) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63123 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K18 Pro protein) (HERV-K110 Pro protein) (HERV-K(C1a) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK9_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.234531 (rank : 18) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
TFP11_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.250389 (rank : 16) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
VPK17_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.230758 (rank : 20) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63125 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.225686 (rank : 25) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63120 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K(C19) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
SMBP2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.104444 (rank : 39) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
TFP11_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.248024 (rank : 17) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
NPHP4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.062024 (rank : 45) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59240 | Gene names | Nphp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-4 (Nephroretinin). | |||||
|
PINX1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.274897 (rank : 14) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
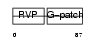
VPK10_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.227956 (rank : 23) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK11_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 28) | NC score | 0.221436 (rank : 27) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63129, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K101 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.227907 (rank : 24) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK5_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.228198 (rank : 22) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 31) | NC score | 0.222699 (rank : 26) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6I0, Q9UKH5 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K(HML-2.HOM) Pro protein) (HERV-K108 Pro protein) (HERV-K(C7) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
PINX1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 32) | NC score | 0.260102 (rank : 15) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
K0082_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.164949 (rank : 30) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
VPK4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.204209 (rank : 29) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63127, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K109 Pro protein) (HERV-K(C6) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.204310 (rank : 28) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63122 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K115 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
FOXJ2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.021781 (rank : 57) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
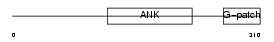
BAT4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.155031 (rank : 32) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
K0082_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.156553 (rank : 31) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
CV106_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.067710 (rank : 44) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P0N0, Q86V14, Q96PY4, Q9NUR5, Q9Y4X9 | Gene names | C14orf106, KIAA1903 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf106 (P243). | |||||
|
ZGPAT_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.151083 (rank : 34) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.018989 (rank : 58) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
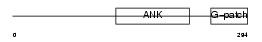
BAT4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.144601 (rank : 36) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
INVO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.013321 (rank : 65) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.038112 (rank : 49) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.147013 (rank : 35) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
NFX1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.030144 (rank : 55) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.031138 (rank : 53) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
GPTC3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.089505 (rank : 40) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
LAMA4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.012478 (rank : 66) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.030226 (rank : 54) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
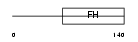
FOXJ2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.016881 (rank : 59) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ES18 | Gene names | Foxj2, Fhx | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
CENG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.010112 (rank : 67) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.014066 (rank : 62) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
E2AK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.004539 (rank : 69) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
WDR60_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.034618 (rank : 51) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.036270 (rank : 50) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.016831 (rank : 60) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.028359 (rank : 56) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CDX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.002888 (rank : 72) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.014056 (rank : 63) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
DSRAD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.045712 (rank : 48) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
CENG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.009037 (rank : 68) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
DSRAD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.031722 (rank : 52) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.003316 (rank : 71) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
MYOTI_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.004017 (rank : 70) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBF9 | Gene names | MYOT, TTID | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotilin (Titin immunoglobulin domain protein) (Myofibrillar titin- like Ig domains protein) (57 kDa cytoskeletal protein). | |||||
|
TANK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.013737 (rank : 64) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70347, Q61178 | Gene names | Tank, Itraf | |||
|
Domain Architecture |

|
|||||
| Description | TRAF family member-associated NF-kappa-B activator (TRAF-interacting protein) (I-TRAF). | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.016697 (rank : 61) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
CN118_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.068800 (rank : 43) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWQ4, Q6PEJ7, Q9NWH0, Q9ULR8 | Gene names | C14orf118, KIAA1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118. | |||||
|
CN118_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.074760 (rank : 42) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PE65, Q6ZPW9, Q8CD08, Q9DA49 | Gene names | Kiaa1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118 homolog. | |||||
|
RBM6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.085196 (rank : 41) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
SPF45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.054450 (rank : 47) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPF45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.056045 (rank : 46) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
NKRF_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
NKRF_HUMAN
|
||||||
| NC score | 0.994796 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
GPTC2_HUMAN
|
||||||
| NC score | 0.397540 (rank : 3) | θ value | 2.61198e-07 (rank : 3) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC2_MOUSE
|
||||||
| NC score | 0.388514 (rank : 4) | θ value | 7.59969e-07 (rank : 6) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
SFR14_HUMAN
|
||||||
| NC score | 0.386732 (rank : 5) | θ value | 3.41135e-07 (rank : 5) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.375008 (rank : 6) | θ value | 2.61198e-07 (rank : 4) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
AGGF1_MOUSE
|
||||||
| NC score | 0.361850 (rank : 7) | θ value | 8.40245e-06 (rank : 7) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
SF04_HUMAN
|
||||||
| NC score | 0.349765 (rank : 8) | θ value | 2.44474e-05 (rank : 8) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SF04_MOUSE
|
||||||
| NC score | 0.342777 (rank : 9) | θ value | 3.19293e-05 (rank : 9) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.340282 (rank : 10) | θ value | 7.1131e-05 (rank : 10) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
RBM5_HUMAN
|
||||||
| NC score | 0.298629 (rank : 11) | θ value | 0.00020696 (rank : 11) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| NC score | 0.298004 (rank : 12) | θ value | 0.00020696 (rank : 12) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
RBM10_HUMAN
|
||||||
| NC score | 0.292343 (rank : 13) | θ value | 0.00134147 (rank : 13) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
PINX1_MOUSE
|
||||||
| NC score | 0.274897 (rank : 14) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
PINX1_HUMAN
|
||||||
| NC score | 0.260102 (rank : 15) | θ value | 0.163984 (rank : 32) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
TFP11_MOUSE
|
||||||
| NC score | 0.250389 (rank : 16) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
TFP11_HUMAN
|
||||||
| NC score | 0.248024 (rank : 17) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
VPK9_HUMAN
|
||||||
| NC score | 0.234531 (rank : 18) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK1_HUMAN
|
||||||
| NC score | 0.233680 (rank : 19) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK17_HUMAN
|
||||||
| NC score | 0.230758 (rank : 20) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63125 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK7_HUMAN
|
||||||
| NC score | 0.229672 (rank : 21) | θ value | 0.0330416 (rank : 18) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63123 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K18 Pro protein) (HERV-K110 Pro protein) (HERV-K(C1a) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK5_HUMAN
|
||||||
| NC score | 0.228198 (rank : 22) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK10_HUMAN
|
||||||
| NC score | 0.227956 (rank : 23) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| NC score | 0.227907 (rank : 24) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK3_HUMAN
|
||||||
| NC score | 0.225686 (rank : 25) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63120 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K(C19) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK2_HUMAN
|
||||||
| NC score | 0.222699 (rank : 26) | θ value | 0.125558 (rank : 31) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6I0, Q9UKH5 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K(HML-2.HOM) Pro protein) (HERV-K108 Pro protein) (HERV-K(C7) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK11_HUMAN
|
||||||
| NC score | 0.221436 (rank : 27) | θ value | 0.0961366 (rank : 28) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63129, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K101 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK6_HUMAN
|
||||||
| NC score | 0.204310 (rank : 28) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63122 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K115 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK4_HUMAN
|
||||||
| NC score | 0.204209 (rank : 29) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63127, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K109 Pro protein) (HERV-K(C6) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
K0082_MOUSE
|
||||||
| NC score | 0.164949 (rank : 30) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
K0082_HUMAN
|
||||||
| NC score | 0.156553 (rank : 31) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
BAT4_MOUSE
|
||||||
| NC score | 0.155031 (rank : 32) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.152779 (rank : 33) | θ value | 0.00298849 (rank : 16) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ZGPAT_MOUSE
|
||||||
| NC score | 0.151083 (rank : 34) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.147013 (rank : 35) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
BAT4_HUMAN
|
||||||
| NC score | 0.144601 (rank : 36) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
SMBP2_MOUSE
|
||||||
| NC score | 0.118952 (rank : 37) | θ value | 0.00228821 (rank : 14) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.106619 (rank : 38) | θ value | 0.00298849 (rank : 15) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.104444 (rank : 39) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
GPTC3_MOUSE
|
||||||
| NC score | 0.089505 (rank : 40) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.085196 (rank : 41) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
CN118_MOUSE
|
||||||
| NC score | 0.074760 (rank : 42) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PE65, Q6ZPW9, Q8CD08, Q9DA49 | Gene names | Kiaa1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118 homolog. | |||||
|
CN118_HUMAN
|
||||||
| NC score | 0.068800 (rank : 43) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWQ4, Q6PEJ7, Q9NWH0, Q9ULR8 | Gene names | C14orf118, KIAA1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118. | |||||
|
CV106_HUMAN
|
||||||
| NC score | 0.067710 (rank : 44) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P0N0, Q86V14, Q96PY4, Q9NUR5, Q9Y4X9 | Gene names | C14orf106, KIAA1903 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf106 (P243). | |||||
|
NPHP4_MOUSE
|
||||||
| NC score | 0.062024 (rank : 45) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59240 | Gene names | Nphp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-4 (Nephroretinin). | |||||
|
SPF45_MOUSE
|
||||||
| NC score | 0.056045 (rank : 46) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPF45_HUMAN
|
||||||
| NC score | 0.054450 (rank : 47) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
DSRAD_MOUSE
|
||||||
| NC score | 0.045712 (rank : 48) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.038112 (rank : 49) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.036270 (rank : 50) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
WDR60_HUMAN
|
||||||
| NC score | 0.034618 (rank : 51) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
DSRAD_HUMAN
|
||||||
| NC score | 0.031722 (rank : 52) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.031138 (rank : 53) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.030226 (rank : 54) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NFX1_HUMAN
|
||||||
| NC score | 0.030144 (rank : 55) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.028359 (rank : 56) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
FOXJ2_HUMAN
|
||||||
| NC score | 0.021781 (rank : 57) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.018989 (rank : 58) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
FOXJ2_MOUSE
|
||||||
| NC score | 0.016881 (rank : 59) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ES18 | Gene names | Foxj2, Fhx | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.016831 (rank : 60) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.016697 (rank : 61) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.014066 (rank : 62) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.014056 (rank : 63) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
TANK_MOUSE
|
||||||
| NC score | 0.013737 (rank : 64) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70347, Q61178 | Gene names | Tank, Itraf | |||
|
Domain Architecture |

|
|||||
| Description | TRAF family member-associated NF-kappa-B activator (TRAF-interacting protein) (I-TRAF). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.013321 (rank : 65) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
LAMA4_HUMAN
|
||||||
| NC score | 0.012478 (rank : 66) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
CENG2_HUMAN
|
||||||
| NC score | 0.010112 (rank : 67) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
CENG2_MOUSE
|
||||||
| NC score | 0.009037 (rank : 68) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
E2AK2_HUMAN
|
||||||
| NC score | 0.004539 (rank : 69) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
MYOTI_HUMAN
|
||||||
| NC score | 0.004017 (rank : 70) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBF9 | Gene names | MYOT, TTID | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotilin (Titin immunoglobulin domain protein) (Myofibrillar titin- like Ig domains protein) (57 kDa cytoskeletal protein). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.003316 (rank : 71) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
CDX1_HUMAN
|
||||||
| NC score | 0.002888 (rank : 72) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||