Please be patient as the page loads
|
CQ028_HUMAN
|
||||||
| SwissProt Accessions | Q8IV36, Q8N5L6, Q8TE83, Q9NT34 | Gene names | C17orf28, DMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 (Down-regulated in multiple cancers- 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CQ028_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IV36, Q8N5L6, Q8TE83, Q9NT34 | Gene names | C17orf28, DMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 (Down-regulated in multiple cancers- 1). | |||||
|
CQ028_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.985465 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R1F6, Q8C8V6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 homolog. | |||||
|
DYM_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 3) | NC score | 0.260233 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHY3 | Gene names | Dym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dymeclin. | |||||
|
DYM_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 4) | NC score | 0.257747 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7RTS9, Q6P2P5, Q8N2M0, Q9BVE9 | Gene names | DYM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dymeclin (Dyggve-Melchior-Clausen syndrome protein). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 5) | NC score | 0.059213 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 6) | NC score | 0.051393 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LIPA4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.063038 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75335, O94971 | Gene names | PPFIA4, KIAA0897 | |||
|
Domain Architecture |
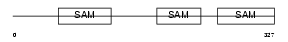
|
|||||
| Description | Liprin-alpha-4 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-4) (PTPRF-interacting protein alpha-4). | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.060900 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.098840 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.053512 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.045057 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
CML2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.017647 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 724 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99527, O00143, O43494, Q13631, Q96F42, Q99981 | Gene names | GPR30, CEPR, CMKRL2, DRY12 | |||
|
Domain Architecture |
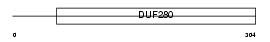
|
|||||
| Description | Chemokine receptor-like 2 (G-protein coupled receptor 30) (IL8-related receptor DRY12) (Flow-induced endothelial G-protein coupled receptor) (FEG-1) (GPCR-BR). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.041663 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
RIPK3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.015169 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |
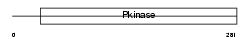
|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.038332 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
PCD12_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.013825 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55134 | Gene names | Pcdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.049808 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
CM007_MOUSE
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.026550 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.036826 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.044005 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.038071 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
REPS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.026028 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.015895 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
DREB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.029325 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.007856 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.022803 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.043463 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.018253 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.021896 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
JIP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.032145 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.023217 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.047775 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.032602 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.016030 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
PAWR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.028738 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
CDN1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.042128 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46527, Q16307, Q9BUS6 | Gene names | CDKN1B, KIP1 | |||
|
Domain Architecture |
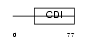
|
|||||
| Description | Cyclin-dependent kinase inhibitor 1B (Cyclin-dependent kinase inhibitor p27) (p27Kip1). | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.030537 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.031319 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.036869 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.044087 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
A4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.011423 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.015946 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
DREB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.035631 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
FRMD6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.010771 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |
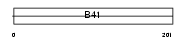
|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.012993 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
LATS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.002101 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRM7, Q9P2X1 | Gene names | LATS2, KPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS2 (EC 2.7.11.1) (Large tumor suppressor homolog 2) (Serine/threonine-protein kinase kpm) (Kinase phosphorylated during mitosis protein) (Warts-like kinase). | |||||
|
PHF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.023121 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
IRAK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.006644 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |
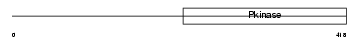
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.035377 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.008325 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.012349 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PSD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.010208 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
RFIP5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.015056 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R361 | Gene names | Rab11fip5, D6Ertd32e, Rip11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11). | |||||
|
SDC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.019165 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75056, Q5T1Z6, Q5T1Z7, Q96CT3, Q96PR8 | Gene names | SDC3, KIAA0468 | |||
|
Domain Architecture |
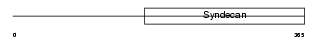
|
|||||
| Description | Syndecan-3 (SYND3). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.008091 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.025524 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
CQ028_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IV36, Q8N5L6, Q8TE83, Q9NT34 | Gene names | C17orf28, DMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 (Down-regulated in multiple cancers- 1). | |||||
|
CQ028_MOUSE
|
||||||
| NC score | 0.985465 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R1F6, Q8C8V6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 homolog. | |||||
|
DYM_MOUSE
|
||||||
| NC score | 0.260233 (rank : 3) | θ value | 4.1701e-05 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHY3 | Gene names | Dym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dymeclin. | |||||
|
DYM_HUMAN
|
||||||
| NC score | 0.257747 (rank : 4) | θ value | 7.1131e-05 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7RTS9, Q6P2P5, Q8N2M0, Q9BVE9 | Gene names | DYM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dymeclin (Dyggve-Melchior-Clausen syndrome protein). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.098840 (rank : 5) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
LIPA4_HUMAN
|
||||||
| NC score | 0.063038 (rank : 6) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75335, O94971 | Gene names | PPFIA4, KIAA0897 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-4 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-4) (PTPRF-interacting protein alpha-4). | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.060900 (rank : 7) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.059213 (rank : 8) | θ value | 0.00298849 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.053512 (rank : 9) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.051393 (rank : 10) | θ value | 0.0252991 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.049808 (rank : 11) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.047775 (rank : 12) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.045057 (rank : 13) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.044087 (rank : 14) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.044005 (rank : 15) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.043463 (rank : 16) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
CDN1B_HUMAN
|
||||||
| NC score | 0.042128 (rank : 17) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46527, Q16307, Q9BUS6 | Gene names | CDKN1B, KIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1B (Cyclin-dependent kinase inhibitor p27) (p27Kip1). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.041663 (rank : 18) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.038332 (rank : 19) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.038071 (rank : 20) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.036869 (rank : 21) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.036826 (rank : 22) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
DREB_MOUSE
|
||||||
| NC score | 0.035631 (rank : 23) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.035377 (rank : 24) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.032602 (rank : 25) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
JIP2_HUMAN
|
||||||
| NC score | 0.032145 (rank : 26) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.031319 (rank : 27) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.030537 (rank : 28) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
DREB_HUMAN
|
||||||
| NC score | 0.029325 (rank : 29) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
PAWR_HUMAN
|
||||||
| NC score | 0.028738 (rank : 30) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
CM007_MOUSE
|
||||||
| NC score | 0.026550 (rank : 31) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
REPS2_HUMAN
|
||||||
| NC score | 0.026028 (rank : 32) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.025524 (rank : 33) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.023217 (rank : 34) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PHF2_HUMAN
|
||||||
| NC score | 0.023121 (rank : 35) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.022803 (rank : 36) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.021896 (rank : 37) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
SDC3_HUMAN
|
||||||
| NC score | 0.019165 (rank : 38) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75056, Q5T1Z6, Q5T1Z7, Q96CT3, Q96PR8 | Gene names | SDC3, KIAA0468 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-3 (SYND3). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.018253 (rank : 39) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
CML2_HUMAN
|
||||||
| NC score | 0.017647 (rank : 40) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 724 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99527, O00143, O43494, Q13631, Q96F42, Q99981 | Gene names | GPR30, CEPR, CMKRL2, DRY12 | |||
|
Domain Architecture |

|
|||||
| Description | Chemokine receptor-like 2 (G-protein coupled receptor 30) (IL8-related receptor DRY12) (Flow-induced endothelial G-protein coupled receptor) (FEG-1) (GPCR-BR). | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.016030 (rank : 41) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.015946 (rank : 42) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.015895 (rank : 43) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
RIPK3_MOUSE
|
||||||
| NC score | 0.015169 (rank : 44) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
RFIP5_MOUSE
|
||||||
| NC score | 0.015056 (rank : 45) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R361 | Gene names | Rab11fip5, D6Ertd32e, Rip11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11). | |||||
|
PCD12_MOUSE
|
||||||
| NC score | 0.013825 (rank : 46) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55134 | Gene names | Pcdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.012993 (rank : 47) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.012349 (rank : 48) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.011423 (rank : 49) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
FRMD6_MOUSE
|
||||||
| NC score | 0.010771 (rank : 50) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
PSD4_HUMAN
|
||||||
| NC score | 0.010208 (rank : 51) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.008325 (rank : 52) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.008091 (rank : 53) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.007856 (rank : 54) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
IRAK1_HUMAN
|
||||||
| NC score | 0.006644 (rank : 55) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
LATS2_HUMAN
|
||||||
| NC score | 0.002101 (rank : 56) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRM7, Q9P2X1 | Gene names | LATS2, KPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS2 (EC 2.7.11.1) (Large tumor suppressor homolog 2) (Serine/threonine-protein kinase kpm) (Kinase phosphorylated during mitosis protein) (Warts-like kinase). | |||||