Please be patient as the page loads
|
RFIP5_MOUSE
|
||||||
| SwissProt Accessions | Q8R361 | Gene names | Rab11fip5, D6Ertd32e, Rip11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RFIP5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.968407 (rank : 2) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXF6, O94939, Q9P0M1 | Gene names | RAB11FIP5, GAF1, KIAA0857, RIP11 | |||
|
Domain Architecture |
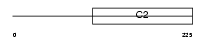
|
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11) (Gamma-SNAP-associated factor 1) (Gaf-1) (Phosphoprotein pp75). | |||||
|
RFIP5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | Q8R361 | Gene names | Rab11fip5, D6Ertd32e, Rip11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11). | |||||
|
RFIP1_MOUSE
|
||||||
| θ value | 2.73936e-73 (rank : 3) | NC score | 0.834955 (rank : 3) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | 4.68348e-65 (rank : 4) | NC score | 0.796747 (rank : 4) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
RFIP2_HUMAN
|
||||||
| θ value | 9.48078e-50 (rank : 5) | NC score | 0.753843 (rank : 5) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 6) | NC score | 0.104105 (rank : 8) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 7) | NC score | 0.101735 (rank : 9) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 8) | NC score | 0.060478 (rank : 15) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 9) | NC score | 0.044223 (rank : 37) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.040841 (rank : 41) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
ASPX_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.072648 (rank : 12) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.067203 (rank : 13) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.047600 (rank : 28) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SDCG1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.046902 (rank : 31) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |
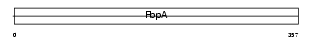
|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
UN13A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.114811 (rank : 7) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.013991 (rank : 99) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
UN13B_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.115867 (rank : 6) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.063466 (rank : 14) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
CD2L5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.007044 (rank : 123) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.050090 (rank : 24) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.036356 (rank : 51) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.052049 (rank : 20) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
DOK7_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.047693 (rank : 27) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
PEPP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.018003 (rank : 86) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHV9, O95030, Q3SYE0 | Gene names | PEPP1, OTEX | |||
|
Domain Architecture |
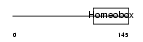
|
|||||
| Description | Paired-like homeobox protein PEPP-1 (Ovary-, testis- and epididymis- expressed gene protein). | |||||
|
RASL2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.074043 (rank : 11) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.051478 (rank : 21) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.057357 (rank : 18) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.015700 (rank : 95) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SREC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.016399 (rank : 92) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
TF3C1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.035538 (rank : 52) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K284, Q8CAL9 | Gene names | Gtf3c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.044740 (rank : 36) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.046988 (rank : 30) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
KIF1C_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.010364 (rank : 113) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43896, O75186 | Gene names | KIF1C, KIAA0706 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
LMO6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.019869 (rank : 80) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80VL3, Q8CAY9 | Gene names | Lmo6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
M6PBP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.028629 (rank : 61) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60664, Q53G77, Q9BS03, Q9UBD7, Q9UP92 | Gene names | M6PRBP1, TIP47 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-6-phosphate receptor-binding protein 1 (Cargo selection protein TIP47) (47 kDa mannose 6-phosphate receptor-binding protein) (47 kDa MPR-binding protein) (Placental protein 17) (PP17). | |||||
|
SCG1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.042200 (rank : 39) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.037003 (rank : 49) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CPSF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.045055 (rank : 34) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10570, Q96AF0 | Gene names | CPSF1, CPSF160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.039900 (rank : 46) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
HLX1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.013618 (rank : 102) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14774, Q15988 | Gene names | HLX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein HLX1 (Homeobox protein HB24). | |||||
|
JHD1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.021000 (rank : 77) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
SSA27_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.042066 (rank : 40) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56873 | Gene names | Sssca1, C184l | |||
|
Domain Architecture |
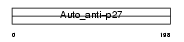
|
|||||
| Description | Sjoegren syndrome/scleroderma autoantigen 1 homolog (Autoantigen p27 homolog) (Protein C184L). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.058338 (rank : 17) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
COJA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.017599 (rank : 87) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
CPSF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.043799 (rank : 38) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPU4 | Gene names | Cpsf1, Cpsf160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.017228 (rank : 88) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.008882 (rank : 117) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
EAN57_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.038051 (rank : 48) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
HXC11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.012198 (rank : 104) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43248, Q96DH2 | Gene names | HOXC11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C11. | |||||
|
MAF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.029128 (rank : 59) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.040520 (rank : 43) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.044838 (rank : 35) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.021381 (rank : 75) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
ABC8B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.007938 (rank : 119) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K440, Q69ZY4, Q8BRQ1, Q9JL38 | Gene names | Abca8b, Abca8, Kiaa0822 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 8-B. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.036980 (rank : 50) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
JUND_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.019045 (rank : 84) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
NFM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.016588 (rank : 91) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NMDE4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.011977 (rank : 105) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
R51A1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.046020 (rank : 33) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.030041 (rank : 56) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TEX2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.040808 (rank : 42) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWB9, Q6AHZ5, Q8N3L0, Q9C0C5 | Gene names | TEX2, KIAA1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
TEX2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.040361 (rank : 44) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.024661 (rank : 68) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.010064 (rank : 114) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.015406 (rank : 96) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.027225 (rank : 62) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.007225 (rank : 121) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.001256 (rank : 131) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.019227 (rank : 83) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
WDR22_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.013677 (rank : 101) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96JK2, O60559, Q8N3V3, Q8N3V5 | Gene names | WDR22, BCRG2, KIAA1824 | |||
|
Domain Architecture |
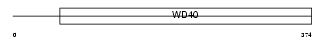
|
|||||
| Description | WD repeat protein 22 (Breakpoint cluster region protein 2) (BCRP2). | |||||
|
AFTIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.022161 (rank : 73) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ULP2, Q6ZM66, Q86VW3, Q8TCF3, Q9H7E3, Q9HAB9, Q9NXS4 | Gene names | AFTPH, AFTH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aftiphilin. | |||||
|
BLNK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.029471 (rank : 58) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
CNKR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.022525 (rank : 71) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
DZIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.016793 (rank : 89) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.030441 (rank : 55) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.022383 (rank : 72) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MAF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.025292 (rank : 66) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
RAB3I_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.026000 (rank : 64) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.011298 (rank : 109) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.018520 (rank : 85) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ACRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.005979 (rank : 125) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10323, Q6ICK2 | Gene names | ACR, ACRS | |||
|
Domain Architecture |
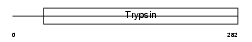
|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
HXD12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.007637 (rank : 120) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23812 | Gene names | Hoxd12, Hox-4.7, Hoxd-12 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D12 (Hox-4.7) (Hox-5.6). | |||||
|
LMO6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.014508 (rank : 98) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |
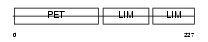
|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
MILK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.016217 (rank : 93) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NFIB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.010584 (rank : 112) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00712, O00166, Q12858, Q96J45 | Gene names | NFIB | |||
|
Domain Architecture |
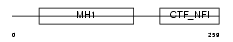
|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.040132 (rank : 45) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.020340 (rank : 78) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.027024 (rank : 63) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.032629 (rank : 54) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.022730 (rank : 70) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.006879 (rank : 124) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.003228 (rank : 127) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.038434 (rank : 47) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.011077 (rank : 111) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
DCX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.019370 (rank : 82) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
FA76B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.011686 (rank : 107) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XP8, Q3UMA6, Q80YR1, Q8CI69 | Gene names | Fam76b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM76B. | |||||
|
FRMD5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.005356 (rank : 126) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
GLI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.001458 (rank : 130) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.034774 (rank : 53) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MAFB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.021136 (rank : 76) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.025846 (rank : 65) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.002126 (rank : 128) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
MEF2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.010020 (rank : 115) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60929 | Gene names | Mef2a | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2A. | |||||
|
NU214_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.028869 (rank : 60) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
OTU7A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.012807 (rank : 103) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.023176 (rank : 69) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
TMUB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.016192 (rank : 94) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JMG3 | Gene names | Tmub1 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1. | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.019590 (rank : 81) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.001686 (rank : 129) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.008227 (rank : 118) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
TTC16_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.013842 (rank : 100) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
ALS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.007074 (rank : 122) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.022062 (rank : 74) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
BLNK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.025244 (rank : 67) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
CN092_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.009486 (rank : 116) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.011530 (rank : 108) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CONA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.011217 (rank : 110) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86Y22, Q8IVR4, Q9NT93 | Gene names | COL23A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
CQ028_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.015056 (rank : 97) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IV36, Q8N5L6, Q8TE83, Q9NT34 | Gene names | C17orf28, DMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 (Down-regulated in multiple cancers- 1). | |||||
|
K0146_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.011851 (rank : 106) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14159, Q96BI5 | Gene names | KIAA0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
PER3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.016780 (rank : 90) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.020158 (rank : 79) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SYT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.049511 (rank : 25) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.049508 (rank : 26) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.046459 (rank : 32) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.047324 (rank : 29) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.029583 (rank : 57) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.050549 (rank : 22) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RASL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.059252 (rank : 16) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RASL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.050545 (rank : 23) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.056759 (rank : 19) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
UN13B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.083863 (rank : 10) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
RFIP5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | Q8R361 | Gene names | Rab11fip5, D6Ertd32e, Rip11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11). | |||||
|
RFIP5_HUMAN
|
||||||
| NC score | 0.968407 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXF6, O94939, Q9P0M1 | Gene names | RAB11FIP5, GAF1, KIAA0857, RIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11) (Gamma-SNAP-associated factor 1) (Gaf-1) (Phosphoprotein pp75). | |||||
|
RFIP1_MOUSE
|
||||||
| NC score | 0.834955 (rank : 3) | θ value | 2.73936e-73 (rank : 3) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.796747 (rank : 4) | θ value | 4.68348e-65 (rank : 4) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
RFIP2_HUMAN
|
||||||
| NC score | 0.753843 (rank : 5) | θ value | 9.48078e-50 (rank : 5) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
UN13B_MOUSE
|
||||||
| NC score | 0.115867 (rank : 6) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13A_HUMAN
|
||||||
| NC score | 0.114811 (rank : 7) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.104105 (rank : 8) | θ value | 0.00134147 (rank : 6) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.101735 (rank : 9) | θ value | 0.00228821 (rank : 7) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
UN13B_HUMAN
|
||||||
| NC score | 0.083863 (rank : 10) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
RASL2_HUMAN
|
||||||
| NC score | 0.074043 (rank : 11) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
ASPX_HUMAN
|
||||||
| NC score | 0.072648 (rank : 12) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.067203 (rank : 13) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.063466 (rank : 14) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.060478 (rank : 15) | θ value | 0.00509761 (rank : 8) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RASL1_HUMAN
|
||||||
| NC score | 0.059252 (rank : 16) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.058338 (rank : 17) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.057357 (rank : 18) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.056759 (rank : 19) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.052049 (rank : 20) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.051478 (rank : 21) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.050549 (rank : 22) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RASL1_MOUSE
|
||||||
| NC score | 0.050545 (rank : 23) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.050090 (rank : 24) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SYT1_HUMAN
|
||||||
| NC score | 0.049511 (rank : 25) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| NC score | 0.049508 (rank : 26) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
DOK7_MOUSE
|
||||||
| NC score | 0.047693 (rank : 27) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.047600 (rank : 28) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SYT2_MOUSE
|
||||||
| NC score | 0.047324 (rank : 29) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.046988 (rank : 30) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
SDCG1_HUMAN
|
||||||
| NC score | 0.046902 (rank : 31) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
SYT2_HUMAN
|
||||||
| NC score | 0.046459 (rank : 32) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.046020 (rank : 33) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
CPSF1_HUMAN
|
||||||
| NC score | 0.045055 (rank : 34) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10570, Q96AF0 | Gene names | CPSF1, CPSF160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.044838 (rank : 35) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.044740 (rank : 36) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.044223 (rank : 37) | θ value | 0.0148317 (rank : 9) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
CPSF1_MOUSE
|
||||||
| NC score | 0.043799 (rank : 38) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPU4 | Gene names | Cpsf1, Cpsf160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
SCG1_HUMAN
|
||||||
| NC score | 0.042200 (rank : 39) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
SSA27_MOUSE
|
||||||
| NC score | 0.042066 (rank : 40) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56873 | Gene names | Sssca1, C184l | |||
|
Domain Architecture |
|
|||||
| Description | Sjoegren syndrome/scleroderma autoantigen 1 homolog (Autoantigen p27 homolog) (Protein C184L). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.040841 (rank : 41) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
TEX2_HUMAN
|
||||||
| NC score | 0.040808 (rank : 42) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWB9, Q6AHZ5, Q8N3L0, Q9C0C5 | Gene names | TEX2, KIAA1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.040520 (rank : 43) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
TEX2_MOUSE
|
||||||
| NC score | 0.040361 (rank : 44) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.040132 (rank : 45) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.039900 (rank : 46) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.038434 (rank : 47) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
EAN57_HUMAN
|
||||||
| NC score | 0.038051 (rank : 48) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.037003 (rank : 49) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.036980 (rank : 50) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.036356 (rank : 51) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
TF3C1_MOUSE
|
||||||
| NC score | 0.035538 (rank : 52) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K284, Q8CAL9 | Gene names | Gtf3c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.034774 (rank : 53) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.032629 (rank : 54) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.030441 (rank : 55) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.030041 (rank : 56) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.029583 (rank : 57) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
BLNK_MOUSE
|
||||||
| NC score | 0.029471 (rank : 58) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
MAF_HUMAN
|
||||||
| NC score | 0.029128 (rank : 59) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.028869 (rank : 60) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
M6PBP_HUMAN
|
||||||
| NC score | 0.028629 (rank : 61) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60664, Q53G77, Q9BS03, Q9UBD7, Q9UP92 | Gene names | M6PRBP1, TIP47 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-6-phosphate receptor-binding protein 1 (Cargo selection protein TIP47) (47 kDa mannose 6-phosphate receptor-binding protein) (47 kDa MPR-binding protein) (Placental protein 17) (PP17). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.027225 (rank : 62) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.027024 (rank : 63) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
RAB3I_HUMAN
|
||||||
| NC score | 0.026000 (rank : 64) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.025846 (rank : 65) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAF_MOUSE
|
||||||
| NC score | 0.025292 (rank : 66) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
BLNK_HUMAN
|
||||||
| NC score | 0.025244 (rank : 67) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.024661 (rank : 68) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.023176 (rank : 69) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.022730 (rank : 70) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
CNKR1_HUMAN
|
||||||
| NC score | 0.022525 (rank : 71) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.022383 (rank : 72) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
AFTIN_HUMAN
|
||||||
| NC score | 0.022161 (rank : 73) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ULP2, Q6ZM66, Q86VW3, Q8TCF3, Q9H7E3, Q9HAB9, Q9NXS4 | Gene names | AFTPH, AFTH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aftiphilin. | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.022062 (rank : 74) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.021381 (rank : 75) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
MAFB_MOUSE
|
||||||
| NC score | 0.021136 (rank : 76) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
JHD1B_MOUSE
|
||||||
| NC score | 0.021000 (rank : 77) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.020340 (rank : 78) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.020158 (rank : 79) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
LMO6_MOUSE
|
||||||
| NC score | 0.019869 (rank : 80) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80VL3, Q8CAY9 | Gene names | Lmo6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.019590 (rank : 81) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.019370 (rank : 82) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.019227 (rank : 83) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
JUND_HUMAN
|
||||||
| NC score | 0.019045 (rank : 84) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.018520 (rank : 85) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
PEPP1_HUMAN
|
||||||
| NC score | 0.018003 (rank : 86) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHV9, O95030, Q3SYE0 | Gene names | PEPP1, OTEX | |||
|
Domain Architecture |

|
|||||
| Description | Paired-like homeobox protein PEPP-1 (Ovary-, testis- and epididymis- expressed gene protein). | |||||
|
COJA1_HUMAN
|
||||||
| NC score | 0.017599 (rank : 87) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.017228 (rank : 88) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
DZIP1_HUMAN
|
||||||
| NC score | 0.016793 (rank : 89) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.016780 (rank : 90) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.016588 (rank : 91) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.016399 (rank : 92) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.016217 (rank : 93) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
TMUB1_MOUSE
|
||||||
| NC score | 0.016192 (rank : 94) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JMG3 | Gene names | Tmub1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1. | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.015700 (rank : 95) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.015406 (rank : 96) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CQ028_HUMAN
|
||||||
| NC score | 0.015056 (rank : 97) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IV36, Q8N5L6, Q8TE83, Q9NT34 | Gene names | C17orf28, DMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 (Down-regulated in multiple cancers- 1). | |||||
|
LMO6_HUMAN
|
||||||
| NC score | 0.014508 (rank : 98) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | 0.013991 (rank : 99) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TTC16_MOUSE
|
||||||
| NC score | 0.013842 (rank : 100) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
WDR22_HUMAN
|
||||||
| NC score | 0.013677 (rank : 101) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96JK2, O60559, Q8N3V3, Q8N3V5 | Gene names | WDR22, BCRG2, KIAA1824 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 22 (Breakpoint cluster region protein 2) (BCRP2). | |||||
|
HLX1_HUMAN
|
||||||
| NC score | 0.013618 (rank : 102) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14774, Q15988 | Gene names | HLX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein HLX1 (Homeobox protein HB24). | |||||
|
OTU7A_HUMAN
|
||||||
| NC score | 0.012807 (rank : 103) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
HXC11_HUMAN
|
||||||
| NC score | 0.012198 (rank : 104) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43248, Q96DH2 | Gene names | HOXC11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C11. | |||||
|
NMDE4_HUMAN
|
||||||
| NC score | 0.011977 (rank : 105) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
K0146_HUMAN
|
||||||
| NC score | 0.011851 (rank : 106) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14159, Q96BI5 | Gene names | KIAA0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
FA76B_MOUSE
|
||||||
| NC score | 0.011686 (rank : 107) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XP8, Q3UMA6, Q80YR1, Q8CI69 | Gene names | Fam76b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM76B. | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.011530 (rank : 108) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.011298 (rank : 109) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
CONA1_HUMAN
|
||||||
| NC score | 0.011217 (rank : 110) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86Y22, Q8IVR4, Q9NT93 | Gene names | COL23A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.011077 (rank : 111) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
NFIB_HUMAN
|
||||||
| NC score | 0.010584 (rank : 112) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00712, O00166, Q12858, Q96J45 | Gene names | NFIB | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
KIF1C_HUMAN
|
||||||
| NC score | 0.010364 (rank : 113) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43896, O75186 | Gene names | KIF1C, KIAA0706 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.010064 (rank : 114) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MEF2A_MOUSE
|
||||||
| NC score | 0.010020 (rank : 115) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60929 | Gene names | Mef2a | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2A. | |||||
|
CN092_HUMAN
|
||||||
| NC score | 0.009486 (rank : 116) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.008882 (rank : 117) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.008227 (rank : 118) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
ABC8B_MOUSE
|
||||||
| NC score | 0.007938 (rank : 119) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K440, Q69ZY4, Q8BRQ1, Q9JL38 | Gene names | Abca8b, Abca8, Kiaa0822 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 8-B. | |||||
|
HXD12_MOUSE
|
||||||
| NC score | 0.007637 (rank : 120) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23812 | Gene names | Hoxd12, Hox-4.7, Hoxd-12 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D12 (Hox-4.7) (Hox-5.6). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.007225 (rank : 121) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.007074 (rank : 122) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
CD2L5_HUMAN
|
||||||
| NC score | 0.007044 (rank : 123) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.006879 (rank : 124) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
ACRO_HUMAN
|
||||||
| NC score | 0.005979 (rank : 125) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10323, Q6ICK2 | Gene names | ACR, ACRS | |||
|
Domain Architecture |

|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
FRMD5_HUMAN
|
||||||
| NC score | 0.005356 (rank : 126) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
ABL1_MOUSE
|
||||||
| NC score | 0.003228 (rank : 127) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
MAST3_HUMAN
|
||||||
| NC score | 0.002126 (rank : 128) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
TRIO_HUMAN
|
||||||
| NC score | 0.001686 (rank : 129) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
GLI1_HUMAN
|
||||||
| NC score | 0.001458 (rank : 130) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
SP2_HUMAN
|
||||||
| NC score | 0.001256 (rank : 131) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 126 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||