Please be patient as the page loads
|
CACB3_MOUSE
|
||||||
| SwissProt Accessions | P54285 | Gene names | Cacnb3, Cacnlb3 | |||
|
Domain Architecture |
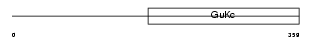
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3) (CCHB3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CACB3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998914 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P54284, Q13913 | Gene names | CACNB3, CACNLB3 | |||
|
Domain Architecture |
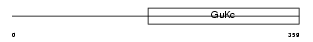
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3). | |||||
|
CACB3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P54285 | Gene names | Cacnb3, Cacnlb3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3) (CCHB3). | |||||
|
CACB4_MOUSE
|
||||||
| θ value | 3.98107e-165 (rank : 3) | NC score | 0.992098 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R0S4, Q8BRN6, Q8CAJ9 | Gene names | Cacnb4, Cacnlb4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
CACB4_HUMAN
|
||||||
| θ value | 6.35588e-163 (rank : 4) | NC score | 0.991162 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00305, O60515, Q96L40 | Gene names | CACNB4, CACNLB4 | |||
|
Domain Architecture |
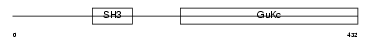
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
CACB1_MOUSE
|
||||||
| θ value | 7.80566e-145 (rank : 5) | NC score | 0.986200 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R3Z5, O88517, Q7TPF2, Q8R3Z6, Q9EPT9 | Gene names | Cacnb1, Cacnlb1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
CACB1_HUMAN
|
||||||
| θ value | 5.05951e-144 (rank : 6) | NC score | 0.986710 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02641, O15331, Q02639, Q02640, Q9C085 | Gene names | CACNB1, CACNLB1 | |||
|
Domain Architecture |
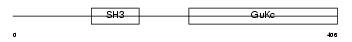
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
CACB2_HUMAN
|
||||||
| θ value | 3.06949e-141 (rank : 7) | NC score | 0.977091 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08289, O00304, Q8WX81, Q96NZ3, Q96NZ4, Q96NZ5, Q9BWU2, Q9HD32, Q9Y340, Q9Y341 | Gene names | CACNB2, CACNLB2, MYSB | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2) (Lambert-Eaton myasthenic syndrome antigen B) (MYSB). | |||||
|
CACB2_MOUSE
|
||||||
| θ value | 1.98959e-140 (rank : 8) | NC score | 0.978521 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CC27, Q8C5J5, Q9CTQ6 | Gene names | Cacnb2, Cacnlb2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2). | |||||
|
DLG1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 9) | NC score | 0.222965 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
DLG3_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 10) | NC score | 0.228795 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92796, Q9ULI8 | Gene names | DLG3, KIAA1232 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102) (Neuroendocrine-DLG) (XLMR). | |||||
|
DLG1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 11) | NC score | 0.222089 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
DLG3_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 12) | NC score | 0.220320 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70175 | Gene names | Dlg3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102). | |||||
|
DLG4_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 13) | NC score | 0.198477 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78352, Q92941, Q9UKK8 | Gene names | DLG4, PSD95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
DLG4_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 14) | NC score | 0.198840 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62108, Q5NCV5, Q5NCV6, Q5NCV7, Q91WJ1 | Gene names | Dlg4, Dlgh4, Psd95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
CSKP_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 15) | NC score | 0.071706 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14936, O43215, Q9NYB3 | Gene names | CASK, LIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (hCASK) (Calcium/calmodulin-dependent serine protein kinase) (Lin-2 homolog). | |||||
|
DLG2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 16) | NC score | 0.195114 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91XM9, Q8BXK7, Q8BYG5 | Gene names | Dlg2, Dlgh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
CSKP_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 17) | NC score | 0.070564 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O70589, O70588 | Gene names | Cask | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (Calcium/calmodulin-dependent serine protein kinase). | |||||
|
MPP6_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.203207 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZW5, Q9H0E1 | Gene names | MPP6, VAM1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Veli-associated MAGUK 1) (VAM-1). | |||||
|
MPP6_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.206453 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLB0, Q9JLB1, Q9WV37 | Gene names | Mpp6, Dlgh4, Pals2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Protein associated with Lin-7 2) (Dlgh4 protein) (P55T protein). | |||||
|
KGUA_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.161479 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64520 | Gene names | Guk1, Gmk | |||
|
Domain Architecture |
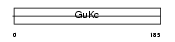
|
|||||
| Description | Guanylate kinase (EC 2.7.4.8) (GMP kinase). | |||||
|
MPP5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.191821 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
DLG2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.182161 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
MPP5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.189713 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLB2 | Gene names | Mpp5, Pals1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5 (Protein associated with Lin-7 1). | |||||
|
NPHP1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.104671 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QY53, Q9D7G8, Q9QZP0, Q9WUZ2 | Gene names | Nphp1, Nph1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1. | |||||
|
EM55_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.177628 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q00013 | Gene names | MPP1, DXS552E, EMP55 | |||
|
Domain Architecture |
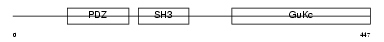
|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1). | |||||
|
EM55_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.175350 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70290 | Gene names | Mpp1 | |||
|
Domain Architecture |
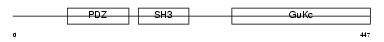
|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1) (Palmitoylated protein p55). | |||||
|
CCD55_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.048751 (rank : 67) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
FOXJ2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.008694 (rank : 79) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
THAP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.020338 (rank : 72) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |
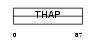
|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
MYO1E_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.012731 (rank : 76) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12965, Q14778 | Gene names | MYO1E, MYO1C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ie (Myosin Ic). | |||||
|
NPHP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.040546 (rank : 68) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15259, O14837 | Gene names | NPHP1, NPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1 (Juvenile nephronophthisis 1 protein). | |||||
|
CD2AP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.021532 (rank : 69) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |
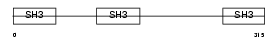
|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
MPP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.129217 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13368 | Gene names | MPP3, DLG3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Discs large homolog 3). | |||||
|
TRPV2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.005967 (rank : 82) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
CITE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.017520 (rank : 73) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99966 | Gene names | CITED1, MSG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cbp/p300-interacting transactivator 1 (Melanocyte-specific protein 1). | |||||
|
HAX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.012359 (rank : 77) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00165, Q96AU4, Q9BS80 | Gene names | HAX1, HS1BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HS1-associating protein X-1 (HAX-1) (HS1-binding protein). | |||||
|
PTK6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.006593 (rank : 80) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q64434 | Gene names | Ptk6, Sik | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase 6 (EC 2.7.10.2) (SRC-related intestinal kinase). | |||||
|
SNW1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.021141 (rank : 70) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13573, Q13483, Q32N03, Q5D0D6 | Gene names | SNW1, SKIIP, SKIP | |||
|
Domain Architecture |
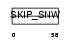
|
|||||
| Description | SNW domain-containing protein 1 (Nuclear protein SkiP) (Ski- interacting protein) (Nuclear receptor coactivator NCoA-62). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.006164 (rank : 81) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.021130 (rank : 71) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
IL6RB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.003838 (rank : 84) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00560 | Gene names | Il6st | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor beta chain precursor (IL-6R-beta) (Interleukin 6 signal transducer) (Membrane glycoprotein 130) (gp130) (CD130 antigen). | |||||
|
MCPH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.011989 (rank : 78) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEM0, Q66GU1, Q9H9C7 | Gene names | MCPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
NDUS8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.014945 (rank : 74) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00217 | Gene names | NDUFS8 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 23 kDa subunit) (Complex I-23kD) (CI-23kD) (TYKY subunit). | |||||
|
NDUS8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.014803 (rank : 75) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K3J1 | Gene names | Ndufs8 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 23 kDa subunit) (Complex I-23kD) (CI-23kD). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.001403 (rank : 86) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.001218 (rank : 87) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PITM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.004396 (rank : 83) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
PTK6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.002699 (rank : 85) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13882 | Gene names | PTK6, BRK | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase 6 (EC 2.7.10.2) (Breast tumor kinase) (Tyrosine-protein kinase BRK). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.066809 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
GRIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.056901 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y3R0 | Gene names | GRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
GRIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.056681 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.055475 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
IL16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.054056 (rank : 55) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
IL16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.051574 (rank : 59) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O54824, O70236, Q3TEM4, Q8R0G5 | Gene names | Il16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
INADL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.055947 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
INADL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.056939 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
KGUA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.133508 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16774 | Gene names | GUK1, GMK | |||
|
Domain Architecture |
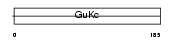
|
|||||
| Description | Guanylate kinase (EC 2.7.4.8) (GMP kinase). | |||||
|
LIN7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.057201 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14910, Q6LES3, Q7LDS4 | Gene names | LIN7A, MALS1, VELI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (hLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein) (Tax interaction protein 33) (TIP-33). | |||||
|
LIN7A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.057172 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8JZS0 | Gene names | Lin7a, Mals1, Veli1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (mLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein). | |||||
|
LIN7B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.057172 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HAP6 | Gene names | LIN7B, MALS2, VELI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (hLin7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein) (hVeli2). | |||||
|
LIN7B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.057078 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88951 | Gene names | Lin7b, Mals2, Veli2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein). | |||||
|
LIN7C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.056637 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NUP9 | Gene names | LIN7C, MALS3, VELI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LIN7C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.056637 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88952 | Gene names | Lin7c, Mals3, Veli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (mLin7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LNX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.051303 (rank : 62) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TBB1, Q8N4C2, Q96MJ7, Q9BY20 | Gene names | LNX1, LNX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
LNX2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.051408 (rank : 60) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XL2, Q8CBE1 | Gene names | Lnx2 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2). | |||||
|
MPDZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.054203 (rank : 54) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
MPDZ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.052763 (rank : 58) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
MPP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.147821 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14168, Q9BQJ2 | Gene names | MPP2, DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Discs large homolog 2). | |||||
|
MPP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.139697 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
MPP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.120517 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88910 | Gene names | Mpp3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Dlgh3 protein). | |||||
|
MPP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.122217 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96JB8, Q6ZNH6, Q96Q43, Q96Q44 | Gene names | MPP4, ALS2CR5, DLG6 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 5 protein). | |||||
|
MPP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.110343 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6P7F1, Q8BTT3, Q8BXM5, Q920P7, Q920P8 | Gene names | Mpp4, Dlg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (MDLG6). | |||||
|
PAR3L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.054850 (rank : 53) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
PARD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.050324 (rank : 66) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
PARD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.053299 (rank : 56) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
PDZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.053190 (rank : 57) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
SYJ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.066080 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P57105, Q96IA4 | Gene names | SYNJ2BP, OMP25 | |||
|
Domain Architecture |
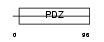
|
|||||
| Description | Synaptojanin-2-binding protein (Mitochondrial outer membrane protein 25). | |||||
|
TS101_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.050481 (rank : 64) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
TX1B3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.051235 (rank : 63) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
TX1B3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.051348 (rank : 61) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
USH1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.050355 (rank : 65) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.066325 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.066182 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.071475 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ZO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.072565 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ZO3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.073340 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
ZO3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.076236 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
CACB3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P54285 | Gene names | Cacnb3, Cacnlb3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3) (CCHB3). | |||||
|
CACB3_HUMAN
|
||||||
| NC score | 0.998914 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P54284, Q13913 | Gene names | CACNB3, CACNLB3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3). | |||||
|
CACB4_MOUSE
|
||||||
| NC score | 0.992098 (rank : 3) | θ value | 3.98107e-165 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R0S4, Q8BRN6, Q8CAJ9 | Gene names | Cacnb4, Cacnlb4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
CACB4_HUMAN
|
||||||
| NC score | 0.991162 (rank : 4) | θ value | 6.35588e-163 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00305, O60515, Q96L40 | Gene names | CACNB4, CACNLB4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
CACB1_HUMAN
|
||||||
| NC score | 0.986710 (rank : 5) | θ value | 5.05951e-144 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02641, O15331, Q02639, Q02640, Q9C085 | Gene names | CACNB1, CACNLB1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
CACB1_MOUSE
|
||||||
| NC score | 0.986200 (rank : 6) | θ value | 7.80566e-145 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R3Z5, O88517, Q7TPF2, Q8R3Z6, Q9EPT9 | Gene names | Cacnb1, Cacnlb1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
CACB2_MOUSE
|
||||||
| NC score | 0.978521 (rank : 7) | θ value | 1.98959e-140 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CC27, Q8C5J5, Q9CTQ6 | Gene names | Cacnb2, Cacnlb2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2). | |||||
|
CACB2_HUMAN
|
||||||
| NC score | 0.977091 (rank : 8) | θ value | 3.06949e-141 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08289, O00304, Q8WX81, Q96NZ3, Q96NZ4, Q96NZ5, Q9BWU2, Q9HD32, Q9Y340, Q9Y341 | Gene names | CACNB2, CACNLB2, MYSB | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2) (Lambert-Eaton myasthenic syndrome antigen B) (MYSB). | |||||
|
DLG3_HUMAN
|
||||||
| NC score | 0.228795 (rank : 9) | θ value | 7.59969e-07 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92796, Q9ULI8 | Gene names | DLG3, KIAA1232 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102) (Neuroendocrine-DLG) (XLMR). | |||||
|
DLG1_HUMAN
|
||||||
| NC score | 0.222965 (rank : 10) | θ value | 4.45536e-07 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
DLG1_MOUSE
|
||||||
| NC score | 0.222089 (rank : 11) | θ value | 2.21117e-06 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
DLG3_MOUSE
|
||||||
| NC score | 0.220320 (rank : 12) | θ value | 0.00020696 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70175 | Gene names | Dlg3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102). | |||||
|
MPP6_MOUSE
|
||||||
| NC score | 0.206453 (rank : 13) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLB0, Q9JLB1, Q9WV37 | Gene names | Mpp6, Dlgh4, Pals2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Protein associated with Lin-7 2) (Dlgh4 protein) (P55T protein). | |||||
|
MPP6_HUMAN
|
||||||
| NC score | 0.203207 (rank : 14) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZW5, Q9H0E1 | Gene names | MPP6, VAM1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Veli-associated MAGUK 1) (VAM-1). | |||||
|
DLG4_MOUSE
|
||||||
| NC score | 0.198840 (rank : 15) | θ value | 0.00035302 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62108, Q5NCV5, Q5NCV6, Q5NCV7, Q91WJ1 | Gene names | Dlg4, Dlgh4, Psd95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
DLG4_HUMAN
|
||||||
| NC score | 0.198477 (rank : 16) | θ value | 0.00035302 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78352, Q92941, Q9UKK8 | Gene names | DLG4, PSD95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
DLG2_MOUSE
|
||||||
| NC score | 0.195114 (rank : 17) | θ value | 0.00175202 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91XM9, Q8BXK7, Q8BYG5 | Gene names | Dlg2, Dlgh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
MPP5_HUMAN
|
||||||
| NC score | 0.191821 (rank : 18) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
MPP5_MOUSE
|
||||||
| NC score | 0.189713 (rank : 19) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLB2 | Gene names | Mpp5, Pals1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5 (Protein associated with Lin-7 1). | |||||
|
DLG2_HUMAN
|
||||||
| NC score | 0.182161 (rank : 20) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
EM55_HUMAN
|
||||||
| NC score | 0.177628 (rank : 21) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q00013 | Gene names | MPP1, DXS552E, EMP55 | |||
|
Domain Architecture |

|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1). | |||||
|
EM55_MOUSE
|
||||||
| NC score | 0.175350 (rank : 22) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70290 | Gene names | Mpp1 | |||
|
Domain Architecture |

|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1) (Palmitoylated protein p55). | |||||
|
KGUA_MOUSE
|
||||||
| NC score | 0.161479 (rank : 23) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64520 | Gene names | Guk1, Gmk | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate kinase (EC 2.7.4.8) (GMP kinase). | |||||
|
MPP2_HUMAN
|
||||||
| NC score | 0.147821 (rank : 24) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14168, Q9BQJ2 | Gene names | MPP2, DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Discs large homolog 2). | |||||
|
MPP2_MOUSE
|
||||||
| NC score | 0.139697 (rank : 25) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
KGUA_HUMAN
|
||||||
| NC score | 0.133508 (rank : 26) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16774 | Gene names | GUK1, GMK | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate kinase (EC 2.7.4.8) (GMP kinase). | |||||
|
MPP3_HUMAN
|
||||||
| NC score | 0.129217 (rank : 27) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13368 | Gene names | MPP3, DLG3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Discs large homolog 3). | |||||
|
MPP4_HUMAN
|
||||||
| NC score | 0.122217 (rank : 28) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96JB8, Q6ZNH6, Q96Q43, Q96Q44 | Gene names | MPP4, ALS2CR5, DLG6 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 5 protein). | |||||
|
MPP3_MOUSE
|
||||||
| NC score | 0.120517 (rank : 29) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88910 | Gene names | Mpp3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Dlgh3 protein). | |||||
|
MPP4_MOUSE
|
||||||
| NC score | 0.110343 (rank : 30) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6P7F1, Q8BTT3, Q8BXM5, Q920P7, Q920P8 | Gene names | Mpp4, Dlg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (MDLG6). | |||||
|
NPHP1_MOUSE
|
||||||
| NC score | 0.104671 (rank : 31) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QY53, Q9D7G8, Q9QZP0, Q9WUZ2 | Gene names | Nphp1, Nph1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1. | |||||
|
ZO3_MOUSE
|
||||||
| NC score | 0.076236 (rank : 32) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
ZO3_HUMAN
|
||||||
| NC score | 0.073340 (rank : 33) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
ZO2_MOUSE
|
||||||
| NC score | 0.072565 (rank : 34) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
CSKP_HUMAN
|
||||||
| NC score | 0.071706 (rank : 35) | θ value | 0.00175202 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14936, O43215, Q9NYB3 | Gene names | CASK, LIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (hCASK) (Calcium/calmodulin-dependent serine protein kinase) (Lin-2 homolog). | |||||
|
ZO2_HUMAN
|
||||||
| NC score | 0.071475 (rank : 36) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
CSKP_MOUSE
|
||||||
| NC score | 0.070564 (rank : 37) | θ value | 0.00298849 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O70589, O70588 | Gene names | Cask | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (Calcium/calmodulin-dependent serine protein kinase). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.066809 (rank : 38) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.066325 (rank : 39) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZO1_MOUSE
|
||||||
| NC score | 0.066182 (rank : 40) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
SYJ2B_HUMAN
|
||||||
| NC score | 0.066080 (rank : 41) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P57105, Q96IA4 | Gene names | SYNJ2BP, OMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2-binding protein (Mitochondrial outer membrane protein 25). | |||||
|
LIN7A_HUMAN
|
||||||
| NC score | 0.057201 (rank : 42) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14910, Q6LES3, Q7LDS4 | Gene names | LIN7A, MALS1, VELI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (hLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein) (Tax interaction protein 33) (TIP-33). | |||||
|
LIN7B_HUMAN
|
||||||
| NC score | 0.057172 (rank : 43) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HAP6 | Gene names | LIN7B, MALS2, VELI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (hLin7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein) (hVeli2). | |||||
|
LIN7A_MOUSE
|
||||||
| NC score | 0.057172 (rank : 44) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8JZS0 | Gene names | Lin7a, Mals1, Veli1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (mLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein). | |||||
|
LIN7B_MOUSE
|
||||||
| NC score | 0.057078 (rank : 45) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88951 | Gene names | Lin7b, Mals2, Veli2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.056939 (rank : 46) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
GRIP1_HUMAN
|
||||||
| NC score | 0.056901 (rank : 47) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y3R0 | Gene names | GRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
GRIP1_MOUSE
|
||||||
| NC score | 0.056681 (rank : 48) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
LIN7C_HUMAN
|
||||||
| NC score | 0.056637 (rank : 49) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NUP9 | Gene names | LIN7C, MALS3, VELI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LIN7C_MOUSE
|
||||||
| NC score | 0.056637 (rank : 50) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88952 | Gene names | Lin7c, Mals3, Veli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (mLin7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.055947 (rank : 51) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.055475 (rank : 52) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
PAR3L_HUMAN
|
||||||
| NC score | 0.054850 (rank : 53) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
MPDZ_HUMAN
|
||||||
| NC score | 0.054203 (rank : 54) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
IL16_HUMAN
|
||||||
| NC score | 0.054056 (rank : 55) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
PARD3_MOUSE
|
||||||
| NC score | 0.053299 (rank : 56) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
PDZD1_HUMAN
|
||||||
| NC score | 0.053190 (rank : 57) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
MPDZ_MOUSE
|
||||||
| NC score | 0.052763 (rank : 58) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
IL16_MOUSE
|
||||||
| NC score | 0.051574 (rank : 59) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O54824, O70236, Q3TEM4, Q8R0G5 | Gene names | Il16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
LNX2_MOUSE
|
||||||
| NC score | 0.051408 (rank : 60) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XL2, Q8CBE1 | Gene names | Lnx2 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2). | |||||
|
TX1B3_MOUSE
|
||||||
| NC score | 0.051348 (rank : 61) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
LNX1_HUMAN
|
||||||
| NC score | 0.051303 (rank : 62) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TBB1, Q8N4C2, Q96MJ7, Q9BY20 | Gene names | LNX1, LNX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
TX1B3_HUMAN
|
||||||
| NC score | 0.051235 (rank : 63) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
TS101_HUMAN
|
||||||
| NC score | 0.050481 (rank : 64) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
USH1C_HUMAN
|
||||||
| NC score | 0.050355 (rank : 65) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
PARD3_HUMAN
|
||||||
| NC score | 0.050324 (rank : 66) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.048751 (rank : 67) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
NPHP1_HUMAN
|
||||||
| NC score | 0.040546 (rank : 68) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15259, O14837 | Gene names | NPHP1, NPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1 (Juvenile nephronophthisis 1 protein). | |||||
|
CD2AP_MOUSE
|
||||||
| NC score | 0.021532 (rank : 69) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |

|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
SNW1_HUMAN
|
||||||
| NC score | 0.021141 (rank : 70) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13573, Q13483, Q32N03, Q5D0D6 | Gene names | SNW1, SKIIP, SKIP | |||
|
Domain Architecture |

|
|||||
| Description | SNW domain-containing protein 1 (Nuclear protein SkiP) (Ski- interacting protein) (Nuclear receptor coactivator NCoA-62). | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.021130 (rank : 71) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP4_HUMAN
|
||||||
| NC score | 0.020338 (rank : 72) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
CITE1_HUMAN
|
||||||
| NC score | 0.017520 (rank : 73) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99966 | Gene names | CITED1, MSG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cbp/p300-interacting transactivator 1 (Melanocyte-specific protein 1). | |||||
|
NDUS8_HUMAN
|
||||||
| NC score | 0.014945 (rank : 74) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00217 | Gene names | NDUFS8 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 23 kDa subunit) (Complex I-23kD) (CI-23kD) (TYKY subunit). | |||||
|
NDUS8_MOUSE
|
||||||
| NC score | 0.014803 (rank : 75) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K3J1 | Gene names | Ndufs8 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 23 kDa subunit) (Complex I-23kD) (CI-23kD). | |||||
|
MYO1E_HUMAN
|
||||||
| NC score | 0.012731 (rank : 76) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12965, Q14778 | Gene names | MYO1E, MYO1C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ie (Myosin Ic). | |||||
|
HAX1_HUMAN
|
||||||
| NC score | 0.012359 (rank : 77) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00165, Q96AU4, Q9BS80 | Gene names | HAX1, HS1BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HS1-associating protein X-1 (HAX-1) (HS1-binding protein). | |||||
|
MCPH1_HUMAN
|
||||||
| NC score | 0.011989 (rank : 78) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEM0, Q66GU1, Q9H9C7 | Gene names | MCPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
FOXJ2_HUMAN
|
||||||
| NC score | 0.008694 (rank : 79) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
PTK6_MOUSE
|
||||||
| NC score | 0.006593 (rank : 80) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q64434 | Gene names | Ptk6, Sik | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase 6 (EC 2.7.10.2) (SRC-related intestinal kinase). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.006164 (rank : 81) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TRPV2_MOUSE
|
||||||
| NC score | 0.005967 (rank : 82) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
PITM2_MOUSE
|
||||||
| NC score | 0.004396 (rank : 83) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
IL6RB_MOUSE
|
||||||
| NC score | 0.003838 (rank : 84) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00560 | Gene names | Il6st | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor beta chain precursor (IL-6R-beta) (Interleukin 6 signal transducer) (Membrane glycoprotein 130) (gp130) (CD130 antigen). | |||||
|
PTK6_HUMAN
|
||||||
| NC score | 0.002699 (rank : 85) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13882 | Gene names | PTK6, BRK | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase 6 (EC 2.7.10.2) (Breast tumor kinase) (Tyrosine-protein kinase BRK). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.001403 (rank : 86) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.001218 (rank : 87) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||