Please be patient as the page loads
|
CACB2_HUMAN
|
||||||
| SwissProt Accessions | Q08289, O00304, Q8WX81, Q96NZ3, Q96NZ4, Q96NZ5, Q9BWU2, Q9HD32, Q9Y340, Q9Y341 | Gene names | CACNB2, CACNLB2, MYSB | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2) (Lambert-Eaton myasthenic syndrome antigen B) (MYSB). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CACB2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q08289, O00304, Q8WX81, Q96NZ3, Q96NZ4, Q96NZ5, Q9BWU2, Q9HD32, Q9Y340, Q9Y341 | Gene names | CACNB2, CACNLB2, MYSB | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2) (Lambert-Eaton myasthenic syndrome antigen B) (MYSB). | |||||
|
CACB2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992828 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8CC27, Q8C5J5, Q9CTQ6 | Gene names | Cacnb2, Cacnlb2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2). | |||||
|
CACB1_MOUSE
|
||||||
| θ value | 7.23842e-175 (rank : 3) | NC score | 0.980118 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R3Z5, O88517, Q7TPF2, Q8R3Z6, Q9EPT9 | Gene names | Cacnb1, Cacnlb1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
CACB1_HUMAN
|
||||||
| θ value | 3.5924e-174 (rank : 4) | NC score | 0.980867 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q02641, O15331, Q02639, Q02640, Q9C085 | Gene names | CACNB1, CACNLB1 | |||
|
Domain Architecture |
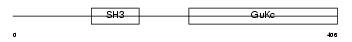
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
CACB4_HUMAN
|
||||||
| θ value | 3.25673e-167 (rank : 5) | NC score | 0.980436 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00305, O60515, Q96L40 | Gene names | CACNB4, CACNLB4 | |||
|
Domain Architecture |
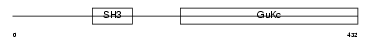
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
CACB4_MOUSE
|
||||||
| θ value | 6.82227e-149 (rank : 6) | NC score | 0.979211 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R0S4, Q8BRN6, Q8CAJ9 | Gene names | Cacnb4, Cacnlb4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
CACB3_HUMAN
|
||||||
| θ value | 1.7995e-141 (rank : 7) | NC score | 0.977246 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54284, Q13913 | Gene names | CACNB3, CACNLB3 | |||
|
Domain Architecture |
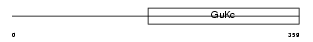
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3). | |||||
|
CACB3_MOUSE
|
||||||
| θ value | 3.06949e-141 (rank : 8) | NC score | 0.977091 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54285 | Gene names | Cacnb3, Cacnlb3 | |||
|
Domain Architecture |
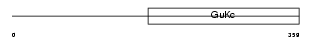
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3) (CCHB3). | |||||
|
DLG3_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 9) | NC score | 0.190173 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70175 | Gene names | Dlg3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102). | |||||
|
DLG3_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 10) | NC score | 0.196271 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92796, Q9ULI8 | Gene names | DLG3, KIAA1232 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102) (Neuroendocrine-DLG) (XLMR). | |||||
|
DLG1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 11) | NC score | 0.189618 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
DLG1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 12) | NC score | 0.190135 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 13) | NC score | 0.101496 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CCD55_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 14) | NC score | 0.099020 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 15) | NC score | 0.092221 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
DLG2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 16) | NC score | 0.152855 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
CSKP_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.061491 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O70589, O70588 | Gene names | Cask | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (Calcium/calmodulin-dependent serine protein kinase). | |||||
|
CSKP_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.062113 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14936, O43215, Q9NYB3 | Gene names | CASK, LIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (hCASK) (Calcium/calmodulin-dependent serine protein kinase) (Lin-2 homolog). | |||||
|
DLG4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.166025 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78352, Q92941, Q9UKK8 | Gene names | DLG4, PSD95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
DLG4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.166377 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62108, Q5NCV5, Q5NCV6, Q5NCV7, Q91WJ1 | Gene names | Dlg4, Dlgh4, Psd95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.040009 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.039678 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
DLG2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.163347 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91XM9, Q8BXK7, Q8BYG5 | Gene names | Dlg2, Dlgh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.043170 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MPP6_MOUSE
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.174820 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JLB0, Q9JLB1, Q9WV37 | Gene names | Mpp6, Dlgh4, Pals2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Protein associated with Lin-7 2) (Dlgh4 protein) (P55T protein). | |||||
|
MPP6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.171760 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZW5, Q9H0E1 | Gene names | MPP6, VAM1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Veli-associated MAGUK 1) (VAM-1). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.030690 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.031643 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
RB15B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.041715 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.042840 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYTL5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.022334 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
EM55_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.148173 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q00013 | Gene names | MPP1, DXS552E, EMP55 | |||
|
Domain Architecture |
|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1). | |||||
|
MPP5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.160766 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
MPP5_MOUSE
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.158860 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLB2 | Gene names | Mpp5, Pals1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5 (Protein associated with Lin-7 1). | |||||
|
TS101_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.075666 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
EM55_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.146360 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70290 | Gene names | Mpp1 | |||
|
Domain Architecture |
|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1) (Palmitoylated protein p55). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.051154 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
RPTN_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.042025 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.008200 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
PRPF3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.036765 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43395, O43446 | Gene names | PRPF3, HPRP3, PRP3 | |||
|
Domain Architecture |
|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3) (U4/U6 snRNP 90 kDa protein) (hPrp3). | |||||
|
SNIP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.052572 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BIZ6, Q3V106, Q8BIZ4 | Gene names | Snip1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.052036 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.039089 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.045164 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.045093 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.030760 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
MARK3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.004673 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.037223 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RB15B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.031177 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.040440 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.012537 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.030363 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
KE4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.020907 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92504, Q5STP8, Q9UIQ0 | Gene names | SLC39A7, HKE4, RING5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc transporter SLC39A7 (Solute carrier family 39 member 7) (Histidine-rich membrane protein Ke4). | |||||
|
MASTL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.005753 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96GX5, Q5T8D5, Q5T8D7, Q8NCD6, Q96SJ5 | Gene names | MASTL, THC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.024904 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.008091 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CCNL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.027451 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.005365 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.036997 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.003710 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.004926 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
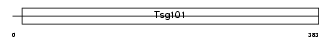
TS101_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.049981 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.017710 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
CD2L5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.006407 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
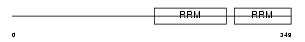
RBM34_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.014822 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42696, Q8N2Z8, Q9H5A1 | Gene names | RBM34, KIAA0117 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 34 (RNA-binding motif protein 34). | |||||
|
SCG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.026503 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.017496 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ARHG4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.020070 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
DDHD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.012053 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.004540 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
MPP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.098696 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96JB8, Q6ZNH6, Q96Q43, Q96Q44 | Gene names | MPP4, ALS2CR5, DLG6 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 5 protein). | |||||
|
PLK4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.003190 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00444, Q8IYF0, Q96Q95 | Gene names | PLK4, SAK, STK18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||
|
PQBP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.033577 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60828, Q4VY25, Q4VY26, Q4VY27, Q4VY29, Q4VY30, Q4VY34, Q4VY35, Q4VY36, Q4VY37, Q4VY38, Q9GZP2, Q9GZU4, Q9GZZ4 | Gene names | PQBP1, NPW38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
PTK6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.006303 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64434 | Gene names | Ptk6, Sik | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase 6 (EC 2.7.10.2) (SRC-related intestinal kinase). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.050678 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.020502 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
ARHG9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.014646 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43307, Q5JSL6 | Gene names | ARHGEF9, KIAA0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin) (PEM-2 homolog). | |||||
|
ARHG9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.014706 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
CCNL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.025770 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
DP13A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.014057 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.014093 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.014042 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.017735 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NPHP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.026928 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15259, O14837 | Gene names | NPHP1, NPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1 (Juvenile nephronophthisis 1 protein). | |||||
|
NPHP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.087326 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QY53, Q9D7G8, Q9QZP0, Q9WUZ2 | Gene names | Nphp1, Nph1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1. | |||||
|
PQBP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.033797 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91VJ5, Q80WW2, Q9ER43, Q9QYY2 | Gene names | Pqbp1, Npw38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.027074 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
BRDT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.010973 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.010327 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.006805 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.003150 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
HXD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.002858 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
HXD3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.002903 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
JHD2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.007785 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
RBY1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.015319 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.021568 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.041296 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.008227 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
TMOD3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.007281 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHJ0, Q3TIT1 | Gene names | Tmod3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-3 (Ubiquitous tropomodulin) (U-Tmod). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.051060 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
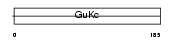
KGUA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.097999 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16774 | Gene names | GUK1, GMK | |||
|
Domain Architecture |
|
|||||
| Description | Guanylate kinase (EC 2.7.4.8) (GMP kinase). | |||||
|
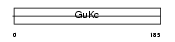
KGUA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.120309 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64520 | Gene names | Guk1, Gmk | |||
|
Domain Architecture |
|
|||||
| Description | Guanylate kinase (EC 2.7.4.8) (GMP kinase). | |||||
|
MPP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.117537 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14168, Q9BQJ2 | Gene names | MPP2, DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Discs large homolog 2). | |||||
|
MPP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.109674 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
MPP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.100742 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13368 | Gene names | MPP3, DLG3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Discs large homolog 3). | |||||
|
MPP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.092924 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88910 | Gene names | Mpp3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Dlgh3 protein). | |||||
|
MPP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.085575 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P7F1, Q8BTT3, Q8BXM5, Q920P7, Q920P8 | Gene names | Mpp4, Dlg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (MDLG6). | |||||
|
SYJ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.050882 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P57105, Q96IA4 | Gene names | SYNJ2BP, OMP25 | |||
|
Domain Architecture |
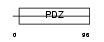
|
|||||
| Description | Synaptojanin-2-binding protein (Mitochondrial outer membrane protein 25). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.051992 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.051558 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.057142 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ZO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.057673 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ZO3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.056966 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
ZO3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.059395 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
CACB2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q08289, O00304, Q8WX81, Q96NZ3, Q96NZ4, Q96NZ5, Q9BWU2, Q9HD32, Q9Y340, Q9Y341 | Gene names | CACNB2, CACNLB2, MYSB | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2) (Lambert-Eaton myasthenic syndrome antigen B) (MYSB). | |||||
|
CACB2_MOUSE
|
||||||
| NC score | 0.992828 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8CC27, Q8C5J5, Q9CTQ6 | Gene names | Cacnb2, Cacnlb2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2). | |||||
|
CACB1_HUMAN
|
||||||
| NC score | 0.980867 (rank : 3) | θ value | 3.5924e-174 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q02641, O15331, Q02639, Q02640, Q9C085 | Gene names | CACNB1, CACNLB1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
CACB4_HUMAN
|
||||||
| NC score | 0.980436 (rank : 4) | θ value | 3.25673e-167 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00305, O60515, Q96L40 | Gene names | CACNB4, CACNLB4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
CACB1_MOUSE
|
||||||
| NC score | 0.980118 (rank : 5) | θ value | 7.23842e-175 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R3Z5, O88517, Q7TPF2, Q8R3Z6, Q9EPT9 | Gene names | Cacnb1, Cacnlb1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
CACB4_MOUSE
|
||||||
| NC score | 0.979211 (rank : 6) | θ value | 6.82227e-149 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R0S4, Q8BRN6, Q8CAJ9 | Gene names | Cacnb4, Cacnlb4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
CACB3_HUMAN
|
||||||
| NC score | 0.977246 (rank : 7) | θ value | 1.7995e-141 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54284, Q13913 | Gene names | CACNB3, CACNLB3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3). | |||||
|
CACB3_MOUSE
|
||||||
| NC score | 0.977091 (rank : 8) | θ value | 3.06949e-141 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54285 | Gene names | Cacnb3, Cacnlb3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3) (CCHB3). | |||||
|
DLG3_HUMAN
|
||||||
| NC score | 0.196271 (rank : 9) | θ value | 0.000270298 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92796, Q9ULI8 | Gene names | DLG3, KIAA1232 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102) (Neuroendocrine-DLG) (XLMR). | |||||
|
DLG3_MOUSE
|
||||||
| NC score | 0.190173 (rank : 10) | θ value | 1.43324e-05 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70175 | Gene names | Dlg3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102). | |||||
|
DLG1_HUMAN
|
||||||
| NC score | 0.190135 (rank : 11) | θ value | 0.000786445 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
DLG1_MOUSE
|
||||||
| NC score | 0.189618 (rank : 12) | θ value | 0.00035302 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
MPP6_MOUSE
|
||||||
| NC score | 0.174820 (rank : 13) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JLB0, Q9JLB1, Q9WV37 | Gene names | Mpp6, Dlgh4, Pals2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Protein associated with Lin-7 2) (Dlgh4 protein) (P55T protein). | |||||
|
MPP6_HUMAN
|
||||||
| NC score | 0.171760 (rank : 14) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZW5, Q9H0E1 | Gene names | MPP6, VAM1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Veli-associated MAGUK 1) (VAM-1). | |||||
|
DLG4_MOUSE
|
||||||
| NC score | 0.166377 (rank : 15) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62108, Q5NCV5, Q5NCV6, Q5NCV7, Q91WJ1 | Gene names | Dlg4, Dlgh4, Psd95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
DLG4_HUMAN
|
||||||
| NC score | 0.166025 (rank : 16) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78352, Q92941, Q9UKK8 | Gene names | DLG4, PSD95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
DLG2_MOUSE
|
||||||
| NC score | 0.163347 (rank : 17) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91XM9, Q8BXK7, Q8BYG5 | Gene names | Dlg2, Dlgh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
MPP5_HUMAN
|
||||||
| NC score | 0.160766 (rank : 18) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
MPP5_MOUSE
|
||||||
| NC score | 0.158860 (rank : 19) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLB2 | Gene names | Mpp5, Pals1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5 (Protein associated with Lin-7 1). | |||||
|
DLG2_HUMAN
|
||||||
| NC score | 0.152855 (rank : 20) | θ value | 0.00869519 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
EM55_HUMAN
|
||||||
| NC score | 0.148173 (rank : 21) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q00013 | Gene names | MPP1, DXS552E, EMP55 | |||
|
Domain Architecture |

|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1). | |||||
|
EM55_MOUSE
|
||||||
| NC score | 0.146360 (rank : 22) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70290 | Gene names | Mpp1 | |||
|
Domain Architecture |

|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1) (Palmitoylated protein p55). | |||||
|
KGUA_MOUSE
|
||||||
| NC score | 0.120309 (rank : 23) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64520 | Gene names | Guk1, Gmk | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate kinase (EC 2.7.4.8) (GMP kinase). | |||||
|
MPP2_HUMAN
|
||||||
| NC score | 0.117537 (rank : 24) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14168, Q9BQJ2 | Gene names | MPP2, DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Discs large homolog 2). | |||||
|
MPP2_MOUSE
|
||||||
| NC score | 0.109674 (rank : 25) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.101496 (rank : 26) | θ value | 0.00298849 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
MPP3_HUMAN
|
||||||
| NC score | 0.100742 (rank : 27) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13368 | Gene names | MPP3, DLG3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Discs large homolog 3). | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.099020 (rank : 28) | θ value | 0.00390308 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
MPP4_HUMAN
|
||||||
| NC score | 0.098696 (rank : 29) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96JB8, Q6ZNH6, Q96Q43, Q96Q44 | Gene names | MPP4, ALS2CR5, DLG6 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 5 protein). | |||||
|
KGUA_HUMAN
|
||||||
| NC score | 0.097999 (rank : 30) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16774 | Gene names | GUK1, GMK | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate kinase (EC 2.7.4.8) (GMP kinase). | |||||
|
MPP3_MOUSE
|
||||||
| NC score | 0.092924 (rank : 31) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88910 | Gene names | Mpp3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Dlgh3 protein). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.092221 (rank : 32) | θ value | 0.00665767 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
NPHP1_MOUSE
|
||||||
| NC score | 0.087326 (rank : 33) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QY53, Q9D7G8, Q9QZP0, Q9WUZ2 | Gene names | Nphp1, Nph1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1. | |||||
|
MPP4_MOUSE
|
||||||
| NC score | 0.085575 (rank : 34) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P7F1, Q8BTT3, Q8BXM5, Q920P7, Q920P8 | Gene names | Mpp4, Dlg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (MDLG6). | |||||
|
TS101_HUMAN
|
||||||
| NC score | 0.075666 (rank : 35) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
CSKP_HUMAN
|
||||||
| NC score | 0.062113 (rank : 36) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14936, O43215, Q9NYB3 | Gene names | CASK, LIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (hCASK) (Calcium/calmodulin-dependent serine protein kinase) (Lin-2 homolog). | |||||
|
CSKP_MOUSE
|
||||||
| NC score | 0.061491 (rank : 37) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O70589, O70588 | Gene names | Cask | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (Calcium/calmodulin-dependent serine protein kinase). | |||||
|
ZO3_MOUSE
|
||||||
| NC score | 0.059395 (rank : 38) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
ZO2_MOUSE
|
||||||
| NC score | 0.057673 (rank : 39) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ZO2_HUMAN
|
||||||
| NC score | 0.057142 (rank : 40) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ZO3_HUMAN
|
||||||
| NC score | 0.056966 (rank : 41) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
SNIP1_MOUSE
|
||||||
| NC score | 0.052572 (rank : 42) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BIZ6, Q3V106, Q8BIZ4 | Gene names | Snip1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.052036 (rank : 43) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.051992 (rank : 44) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZO1_MOUSE
|
||||||
| NC score | 0.051558 (rank : 45) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.051154 (rank : 46) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.051060 (rank : 47) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
SYJ2B_HUMAN
|
||||||
| NC score | 0.050882 (rank : 48) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P57105, Q96IA4 | Gene names | SYNJ2BP, OMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2-binding protein (Mitochondrial outer membrane protein 25). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.050678 (rank : 49) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
TS101_MOUSE
|
||||||
| NC score | 0.049981 (rank : 50) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.045164 (rank : 51) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.045093 (rank : 52) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.043170 (rank : 53) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.042840 (rank : 54) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.042025 (rank : 55) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
RB15B_HUMAN
|
||||||
| NC score | 0.041715 (rank : 56) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.041296 (rank : 57) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.040440 (rank : 58) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.040009 (rank : 59) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.039678 (rank : 60) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.039089 (rank : 61) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.037223 (rank : 62) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.036997 (rank : 63) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
PRPF3_HUMAN
|
||||||
| NC score | 0.036765 (rank : 64) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43395, O43446 | Gene names | PRPF3, HPRP3, PRP3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3) (U4/U6 snRNP 90 kDa protein) (hPrp3). | |||||
|
PQBP1_MOUSE
|
||||||
| NC score | 0.033797 (rank : 65) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91VJ5, Q80WW2, Q9ER43, Q9QYY2 | Gene names | Pqbp1, Npw38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
PQBP1_HUMAN
|
||||||
| NC score | 0.033577 (rank : 66) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60828, Q4VY25, Q4VY26, Q4VY27, Q4VY29, Q4VY30, Q4VY34, Q4VY35, Q4VY36, Q4VY37, Q4VY38, Q9GZP2, Q9GZU4, Q9GZZ4 | Gene names | PQBP1, NPW38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.031643 (rank : 67) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
RB15B_MOUSE
|
||||||
| NC score | 0.031177 (rank : 68) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.030760 (rank : 69) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.030690 (rank : 70) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.030363 (rank : 71) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
CCNL1_MOUSE
|
||||||
| NC score | 0.027451 (rank : 72) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.027074 (rank : 73) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
NPHP1_HUMAN
|
||||||
| NC score | 0.026928 (rank : 74) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15259, O14837 | Gene names | NPHP1, NPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1 (Juvenile nephronophthisis 1 protein). | |||||
|
SCG1_MOUSE
|
||||||
| NC score | 0.026503 (rank : 75) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
CCNL1_HUMAN
|
||||||
| NC score | 0.025770 (rank : 76) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.024904 (rank : 77) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
SYTL5_MOUSE
|
||||||
| NC score | 0.022334 (rank : 78) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.021568 (rank : 79) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
KE4_HUMAN
|
||||||
| NC score | 0.020907 (rank : 80) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92504, Q5STP8, Q9UIQ0 | Gene names | SLC39A7, HKE4, RING5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc transporter SLC39A7 (Solute carrier family 39 member 7) (Histidine-rich membrane protein Ke4). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.020502 (rank : 81) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
ARHG4_HUMAN
|
||||||
| NC score | 0.020070 (rank : 82) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.017735 (rank : 83) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.017710 (rank : 84) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.017496 (rank : 85) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
RBY1A_HUMAN
|
||||||
| NC score | 0.015319 (rank : 86) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
RBM34_HUMAN
|
||||||
| NC score | 0.014822 (rank : 87) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42696, Q8N2Z8, Q9H5A1 | Gene names | RBM34, KIAA0117 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 34 (RNA-binding motif protein 34). | |||||
|
ARHG9_MOUSE
|
||||||
| NC score | 0.014706 (rank : 88) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
ARHG9_HUMAN
|
||||||
| NC score | 0.014646 (rank : 89) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43307, Q5JSL6 | Gene names | ARHGEF9, KIAA0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin) (PEM-2 homolog). | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.014093 (rank : 90) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_HUMAN
|
||||||
| NC score | 0.014057 (rank : 91) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.014042 (rank : 92) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.012537 (rank : 93) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
DDHD1_HUMAN
|
||||||
| NC score | 0.012053 (rank : 94) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.010973 (rank : 95) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.010327 (rank : 96) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.008227 (rank : 97) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.008200 (rank : 98) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.008091 (rank : 99) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
JHD2B_HUMAN
|
||||||
| NC score | 0.007785 (rank : 100) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
TMOD3_MOUSE
|
||||||
| NC score | 0.007281 (rank : 101) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHJ0, Q3TIT1 | Gene names | Tmod3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-3 (Ubiquitous tropomodulin) (U-Tmod). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.006805 (rank : 102) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
CD2L5_HUMAN
|
||||||
| NC score | 0.006407 (rank : 103) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
PTK6_MOUSE
|
||||||
| NC score | 0.006303 (rank : 104) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64434 | Gene names | Ptk6, Sik | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase 6 (EC 2.7.10.2) (SRC-related intestinal kinase). | |||||
|
MASTL_HUMAN
|
||||||
| NC score | 0.005753 (rank : 105) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96GX5, Q5T8D5, Q5T8D7, Q8NCD6, Q96SJ5 | Gene names | MASTL, THC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | 0.005365 (rank : 106) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.004926 (rank : 107) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
MARK3_HUMAN
|
||||||
| NC score | 0.004673 (rank : 108) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.004540 (rank : 109) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.003710 (rank : 110) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
PLK4_HUMAN
|
||||||
| NC score | 0.003190 (rank : 111) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00444, Q8IYF0, Q96Q95 | Gene names | PLK4, SAK, STK18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | 0.003150 (rank : 112) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
HXD3_MOUSE
|
||||||
| NC score | 0.002903 (rank : 113) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
HXD3_HUMAN
|
||||||
| NC score | 0.002858 (rank : 114) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||