Please be patient as the page loads
|
S12A2_HUMAN
|
||||||
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
S12A1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992813 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13621 | Gene names | SLC12A1, NKCC2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992933 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55014, O35938, Q91W55, Q9Z1E0 | Gene names | Slc12a1, Nkcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (BSC1) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.996600 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.989697 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P55017 | Gene names | SLC12A3, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.986760 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59158 | Gene names | Slc12a3, Tsc | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A7_MOUSE
|
||||||
| θ value | 3.84025e-59 (rank : 7) | NC score | 0.815241 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A7_HUMAN
|
||||||
| θ value | 2.10721e-57 (rank : 8) | NC score | 0.818451 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y666, Q96I81, Q9H7I3, Q9H7I7, Q9UFW2 | Gene names | SLC12A7, KCC4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A4_HUMAN
|
||||||
| θ value | 1.36585e-56 (rank : 9) | NC score | 0.810434 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UP95, O60632, O75893, Q13953, Q96LD5 | Gene names | SLC12A4, KCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (hKCC1). | |||||
|
S12A4_MOUSE
|
||||||
| θ value | 1.36585e-56 (rank : 10) | NC score | 0.811313 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JIS8, O55069, Q9ET57 | Gene names | Slc12a4, Kcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (mKCC1). | |||||
|
S12A5_HUMAN
|
||||||
| θ value | 5.73848e-55 (rank : 11) | NC score | 0.811584 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
S12A5_MOUSE
|
||||||
| θ value | 9.78833e-55 (rank : 12) | NC score | 0.811500 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
S12A6_HUMAN
|
||||||
| θ value | 2.18062e-54 (rank : 13) | NC score | 0.807786 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| θ value | 1.08223e-53 (rank : 14) | NC score | 0.806912 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
AAA1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 15) | NC score | 0.087561 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P63115, Q9CW25, Q9JMH8 | Gene names | Slc7a10, Asc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1) (D-serine transporter). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.017195 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
CTR2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.154582 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
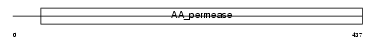
CTR2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.140185 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52569 | Gene names | SLC7A2, ATRC2 | |||
|
Domain Architecture |
|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2). | |||||
|
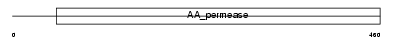
LAT2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.079273 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXW9 | Gene names | Slc7a8, Lat2 | |||
|
Domain Architecture |
|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2). | |||||
|
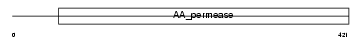
CTR1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.138506 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
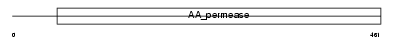
LAT2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.076791 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UHI5, Q9UKQ6, Q9UKQ7, Q9UKQ8, Q9Y445 | Gene names | SLC7A8, LAT2 | |||
|
Domain Architecture |
|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2) (hLAT2). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.010787 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
GSDM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.016649 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QA5, Q8N1M6 | Gene names | GSDM1, GSDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gasdermin. | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.015190 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.026352 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CLMN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.011440 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
NEST_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.007565 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.004016 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
FOXL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.004453 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12952, Q9H242 | Gene names | FOXL1, FKHL11, FREAC7 | |||
|
Domain Architecture |
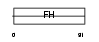
|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Forkhead- related transcription factor 7) (FREAC-7). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.010132 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
PRAX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.012396 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
AAA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.054199 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NS82 | Gene names | SLC7A10, ASC1 | |||
|
Domain Architecture |
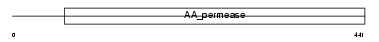
|
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1). | |||||
|
HOOK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.005149 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.003761 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KIF6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.003559 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZMV9, Q2MDE3, Q2MDE4, Q5T8J6, Q6ZWE3, Q86T87, Q8WTV4 | Gene names | KIF6, C6orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF6. | |||||
|
KLF14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | -0.001202 (rank : 60) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TD94 | Gene names | KLF14, BTEB5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 14 (Transcription factor BTEB5) (Basic transcription element-binding protein 5) (BTE-binding protein 5). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.002678 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.002718 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.005731 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.007313 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
CLIC6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.007905 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CTR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.103134 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |
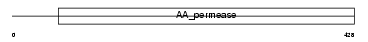
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.002523 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
GLI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | -0.001487 (rank : 62) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.012028 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.004526 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.006897 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.005671 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
EGLN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.005615 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.003935 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
OLF15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | -0.000753 (rank : 59) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23275 | Gene names | Olfr15 | |||
|
Domain Architecture |
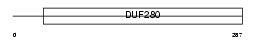
|
|||||
| Description | Olfactory receptor 15 (Odorant receptor OR3). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.004155 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | -0.001309 (rank : 61) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SSFA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.009071 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28290, Q96E36 | Gene names | SSFA2, CS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-specific antigen 2 (Cleavage signal-1 protein) (CS-1). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.006957 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.006608 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
BAT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.059289 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P82251 | Gene names | SLC7A9, BAT1 | |||
|
Domain Architecture |
|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
BAT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.069562 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXA6 | Gene names | Slc7a9, Bat1 | |||
|
Domain Architecture |
|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
CTR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.075756 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |
|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
CTR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.093303 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
CTR4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.052102 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43246, Q3KNQ6, Q4VC45, Q6IBY8, Q96H88 | Gene names | SLC7A4 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 4 (CAT-4) (CAT4). | |||||
|
XCT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.057811 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WTR6 | Gene names | Slc7a11 | |||
|
Domain Architecture |
|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT). | |||||
|
S12A2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A2_MOUSE
|
||||||
| NC score | 0.996600 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A1_MOUSE
|
||||||
| NC score | 0.992933 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55014, O35938, Q91W55, Q9Z1E0 | Gene names | Slc12a1, Nkcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (BSC1) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A1_HUMAN
|
||||||
| NC score | 0.992813 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13621 | Gene names | SLC12A1, NKCC2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A3_HUMAN
|
||||||
| NC score | 0.989697 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P55017 | Gene names | SLC12A3, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A3_MOUSE
|
||||||
| NC score | 0.986760 (rank : 6) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59158 | Gene names | Slc12a3, Tsc | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A7_HUMAN
|
||||||
| NC score | 0.818451 (rank : 7) | θ value | 2.10721e-57 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y666, Q96I81, Q9H7I3, Q9H7I7, Q9UFW2 | Gene names | SLC12A7, KCC4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A7_MOUSE
|
||||||
| NC score | 0.815241 (rank : 8) | θ value | 3.84025e-59 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A5_HUMAN
|
||||||
| NC score | 0.811584 (rank : 9) | θ value | 5.73848e-55 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
S12A5_MOUSE
|
||||||
| NC score | 0.811500 (rank : 10) | θ value | 9.78833e-55 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
S12A4_MOUSE
|
||||||
| NC score | 0.811313 (rank : 11) | θ value | 1.36585e-56 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JIS8, O55069, Q9ET57 | Gene names | Slc12a4, Kcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (mKCC1). | |||||
|
S12A4_HUMAN
|
||||||
| NC score | 0.810434 (rank : 12) | θ value | 1.36585e-56 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UP95, O60632, O75893, Q13953, Q96LD5 | Gene names | SLC12A4, KCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (hKCC1). | |||||
|
S12A6_HUMAN
|
||||||
| NC score | 0.807786 (rank : 13) | θ value | 2.18062e-54 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| NC score | 0.806912 (rank : 14) | θ value | 1.08223e-53 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
CTR2_MOUSE
|
||||||
| NC score | 0.154582 (rank : 15) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
CTR2_HUMAN
|
||||||
| NC score | 0.140185 (rank : 16) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52569 | Gene names | SLC7A2, ATRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2). | |||||
|
CTR1_MOUSE
|
||||||
| NC score | 0.138506 (rank : 17) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
CTR1_HUMAN
|
||||||
| NC score | 0.103134 (rank : 18) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
CTR3_MOUSE
|
||||||
| NC score | 0.093303 (rank : 19) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
AAA1_MOUSE
|
||||||
| NC score | 0.087561 (rank : 20) | θ value | 0.0148317 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P63115, Q9CW25, Q9JMH8 | Gene names | Slc7a10, Asc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1) (D-serine transporter). | |||||
|
LAT2_MOUSE
|
||||||
| NC score | 0.079273 (rank : 21) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXW9 | Gene names | Slc7a8, Lat2 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2). | |||||
|
LAT2_HUMAN
|
||||||
| NC score | 0.076791 (rank : 22) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UHI5, Q9UKQ6, Q9UKQ7, Q9UKQ8, Q9Y445 | Gene names | SLC7A8, LAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2) (hLAT2). | |||||
|
CTR3_HUMAN
|
||||||
| NC score | 0.075756 (rank : 23) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
BAT1_MOUSE
|
||||||
| NC score | 0.069562 (rank : 24) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXA6 | Gene names | Slc7a9, Bat1 | |||
|
Domain Architecture |

|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
BAT1_HUMAN
|
||||||
| NC score | 0.059289 (rank : 25) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P82251 | Gene names | SLC7A9, BAT1 | |||
|
Domain Architecture |

|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
XCT_MOUSE
|
||||||
| NC score | 0.057811 (rank : 26) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WTR6 | Gene names | Slc7a11 | |||
|
Domain Architecture |

|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT). | |||||
|
AAA1_HUMAN
|
||||||
| NC score | 0.054199 (rank : 27) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NS82 | Gene names | SLC7A10, ASC1 | |||
|
Domain Architecture |

|
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1). | |||||
|
CTR4_HUMAN
|
||||||
| NC score | 0.052102 (rank : 28) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43246, Q3KNQ6, Q4VC45, Q6IBY8, Q96H88 | Gene names | SLC7A4 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 4 (CAT-4) (CAT4). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.026352 (rank : 29) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.017195 (rank : 30) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
GSDM1_HUMAN
|
||||||
| NC score | 0.016649 (rank : 31) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QA5, Q8N1M6 | Gene names | GSDM1, GSDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gasdermin. | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.015190 (rank : 32) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.012396 (rank : 33) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.012028 (rank : 34) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
CLMN_MOUSE
|
||||||
| NC score | 0.011440 (rank : 35) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.010787 (rank : 36) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.010132 (rank : 37) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
SSFA2_HUMAN
|
||||||
| NC score | 0.009071 (rank : 38) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28290, Q96E36 | Gene names | SSFA2, CS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-specific antigen 2 (Cleavage signal-1 protein) (CS-1). | |||||
|
CLIC6_MOUSE
|
||||||
| NC score | 0.007905 (rank : 39) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.007565 (rank : 40) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.007313 (rank : 41) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.006957 (rank : 42) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.006897 (rank : 43) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.006608 (rank : 44) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.005731 (rank : 45) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.005671 (rank : 46) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
EGLN1_MOUSE
|
||||||
| NC score | 0.005615 (rank : 47) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
HOOK1_MOUSE
|
||||||
| NC score | 0.005149 (rank : 48) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.004526 (rank : 49) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
FOXL1_HUMAN
|
||||||
| NC score | 0.004453 (rank : 50) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12952, Q9H242 | Gene names | FOXL1, FKHL11, FREAC7 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Forkhead- related transcription factor 7) (FREAC-7). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.004155 (rank : 51) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.004016 (rank : 52) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.003935 (rank : 53) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.003761 (rank : 54) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KIF6_HUMAN
|
||||||
| NC score | 0.003559 (rank : 55) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZMV9, Q2MDE3, Q2MDE4, Q5T8J6, Q6ZWE3, Q86T87, Q8WTV4 | Gene names | KIF6, C6orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF6. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.002718 (rank : 56) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.002678 (rank : 57) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.002523 (rank : 58) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
OLF15_MOUSE
|
||||||
| NC score | -0.000753 (rank : 59) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23275 | Gene names | Olfr15 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 15 (Odorant receptor OR3). | |||||
|
KLF14_HUMAN
|
||||||
| NC score | -0.001202 (rank : 60) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TD94 | Gene names | KLF14, BTEB5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 14 (Transcription factor BTEB5) (Basic transcription element-binding protein 5) (BTE-binding protein 5). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | -0.001309 (rank : 61) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
GLI1_HUMAN
|
||||||
| NC score | -0.001487 (rank : 62) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||