Please be patient as the page loads
|
CAPS2_MOUSE
|
||||||
| SwissProt Accessions | Q8BYR5, O08903, Q66JM7, Q6PCL7, Q76I88, Q7TMM6, Q80ZV8, Q8BL25, Q8BY04, Q8K3K6 | Gene names | Cadps2, Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAPS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.983724 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULU8, Q13339, Q6GQQ6, Q8N2Z5, Q8N3M7, Q8NFR0, Q96BC2 | Gene names | CADPS, CAPS, CAPS1, KIAA1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CAPS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.983125 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80TJ1, Q3TSP2, Q61374, Q6AXB4, Q6PGF0 | Gene names | Cadps, Caps, Caps1, Kiaa1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CAPS2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.997268 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q86UW7, Q658Q2, Q7Z5T7, Q8IZW9, Q8N7M4, Q9H6P4, Q9HCI1, Q9NWK8 | Gene names | CADPS2, CAPS2, KIAA1591 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
CAPS2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BYR5, O08903, Q66JM7, Q6PCL7, Q76I88, Q7TMM6, Q80ZV8, Q8BL25, Q8BY04, Q8K3K6 | Gene names | Cadps2, Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
UN13B_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 5) | NC score | 0.181965 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13B_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 6) | NC score | 0.174352 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13A_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 7) | NC score | 0.176604 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 8) | NC score | 0.056122 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.050475 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.076472 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.066484 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.102094 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.038089 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CENB2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.062070 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.053411 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.098507 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
SEP10_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.031868 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C650, Q7TSA5 | Gene names | Sept10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.037104 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
ARBK1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.009951 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P25098, Q13837 | Gene names | ADRBK1, BARK, BARK1, GRK2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adrenergic receptor kinase 1 (EC 2.7.11.15) (Beta-ARK-1) (G- protein coupled receptor kinase 2). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.061958 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.040121 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.094678 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.040660 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.034202 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
SEP10_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.028922 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P0V9, Q9HAH6 | Gene names | SEPT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
UBP7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.028716 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
FKRP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.061065 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H9S5 | Gene names | FKRP | |||
|
Domain Architecture |

|
|||||
| Description | Fukutin-related protein (EC 2.-.-.-). | |||||
|
NEMO_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.037387 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.020329 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
ARBK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.008004 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99MK8, Q99LL8 | Gene names | Adrbk1, Grk2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adrenergic receptor kinase 1 (EC 2.7.11.15) (Beta-ARK-1) (G- protein-coupled receptor kinase 2). | |||||
|
FKRP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.058590 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG64, Q8BJR3 | Gene names | Fkrp | |||
|
Domain Architecture |

|
|||||
| Description | Fukutin-related protein (EC 2.-.-.-). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.057999 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
ANR35_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.020366 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.024283 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
CYH4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.052837 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.020410 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.050815 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.033254 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.048595 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.046129 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.084329 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PRAX_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.041472 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.035053 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.038389 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.029776 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.087434 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.053513 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
CYH3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.045978 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
CYH4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.041388 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
GRB7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.015278 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14451, Q92568, Q96DF9 | Gene names | GRB7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth factor receptor-bound protein 7 (GRB7 adapter protein) (Epidermal growth factor receptor GRB-7) (B47). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.026301 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.015882 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.015831 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CYH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.046127 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.046131 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
EYA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.015617 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |
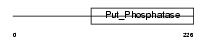
|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.043343 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.031103 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.028398 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
PHLB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.035910 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
SACS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.011821 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
ARAF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.003650 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||
|
CENA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.020536 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.024475 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CYH3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.051975 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
KNTC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.041674 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.019742 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.028397 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.027671 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.027858 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.010271 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CYH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.038849 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.038873 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |
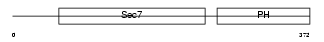
|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.019887 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
MORN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.008677 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
MYO6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.020904 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.030885 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.025408 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CAPS2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BYR5, O08903, Q66JM7, Q6PCL7, Q76I88, Q7TMM6, Q80ZV8, Q8BL25, Q8BY04, Q8K3K6 | Gene names | Cadps2, Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
CAPS2_HUMAN
|
||||||
| NC score | 0.997268 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q86UW7, Q658Q2, Q7Z5T7, Q8IZW9, Q8N7M4, Q9H6P4, Q9HCI1, Q9NWK8 | Gene names | CADPS2, CAPS2, KIAA1591 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
CAPS1_HUMAN
|
||||||
| NC score | 0.983724 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULU8, Q13339, Q6GQQ6, Q8N2Z5, Q8N3M7, Q8NFR0, Q96BC2 | Gene names | CADPS, CAPS, CAPS1, KIAA1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CAPS1_MOUSE
|
||||||
| NC score | 0.983125 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80TJ1, Q3TSP2, Q61374, Q6AXB4, Q6PGF0 | Gene names | Cadps, Caps, Caps1, Kiaa1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
UN13B_HUMAN
|
||||||
| NC score | 0.181965 (rank : 5) | θ value | 2.61198e-07 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13A_HUMAN
|
||||||
| NC score | 0.176604 (rank : 6) | θ value | 3.77169e-06 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
UN13B_MOUSE
|
||||||
| NC score | 0.174352 (rank : 7) | θ value | 7.59969e-07 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.102094 (rank : 8) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.098507 (rank : 9) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.094678 (rank : 10) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.087434 (rank : 11) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.084329 (rank : 12) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.076472 (rank : 13) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.066484 (rank : 14) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.062070 (rank : 15) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.061958 (rank : 16) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
FKRP_HUMAN
|
||||||
| NC score | 0.061065 (rank : 17) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H9S5 | Gene names | FKRP | |||
|
Domain Architecture |

|
|||||
| Description | Fukutin-related protein (EC 2.-.-.-). | |||||
|
FKRP_MOUSE
|
||||||
| NC score | 0.058590 (rank : 18) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG64, Q8BJR3 | Gene names | Fkrp | |||
|
Domain Architecture |

|
|||||
| Description | Fukutin-related protein (EC 2.-.-.-). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.057999 (rank : 19) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.056122 (rank : 20) | θ value | 0.00298849 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.053513 (rank : 21) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.053411 (rank : 22) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CYH4_HUMAN
|
||||||
| NC score | 0.052837 (rank : 23) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
CYH3_HUMAN
|
||||||
| NC score | 0.051975 (rank : 24) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.050815 (rank : 25) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.050475 (rank : 26) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.048595 (rank : 27) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CYH2_MOUSE
|
||||||
| NC score | 0.046131 (rank : 28) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.046129 (rank : 29) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
CYH2_HUMAN
|
||||||
| NC score | 0.046127 (rank : 30) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH3_MOUSE
|
||||||
| NC score | 0.045978 (rank : 31) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.043343 (rank : 32) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
KNTC2_HUMAN
|
||||||
| NC score | 0.041674 (rank : 33) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
PRAX_MOUSE
|
||||||
| NC score | 0.041472 (rank : 34) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
CYH4_MOUSE
|
||||||
| NC score | 0.041388 (rank : 35) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.040660 (rank : 36) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.040121 (rank : 37) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
CYH1_MOUSE
|
||||||
| NC score | 0.038873 (rank : 38) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH1_HUMAN
|
||||||
| NC score | 0.038849 (rank : 39) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.038389 (rank : 40) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.038089 (rank : 41) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
NEMO_MOUSE
|
||||||
| NC score | 0.037387 (rank : 42) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.037104 (rank : 43) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
PHLB1_HUMAN
|
||||||
| NC score | 0.035910 (rank : 44) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.035053 (rank : 45) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.034202 (rank : 46) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.033254 (rank : 47) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
SEP10_MOUSE
|
||||||
| NC score | 0.031868 (rank : 48) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C650, Q7TSA5 | Gene names | Sept10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.031103 (rank : 49) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.030885 (rank : 50) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.029776 (rank : 51) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
SEP10_HUMAN
|
||||||
| NC score | 0.028922 (rank : 52) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P0V9, Q9HAH6 | Gene names | SEPT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
UBP7_HUMAN
|
||||||
| NC score | 0.028716 (rank : 53) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.028398 (rank : 54) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.028397 (rank : 55) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.027858 (rank : 56) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.027671 (rank : 57) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.026301 (rank : 58) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.025408 (rank : 59) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.024475 (rank : 60) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.024283 (rank : 61) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
MYO6_MOUSE
|
||||||
| NC score | 0.020904 (rank : 62) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
CENA1_HUMAN
|
||||||
| NC score | 0.020536 (rank : 63) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.020410 (rank : 64) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.020366 (rank : 65) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.020329 (rank : 66) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.019887 (rank : 67) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.019742 (rank : 68) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.015882 (rank : 69) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.015831 (rank : 70) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
EYA2_HUMAN
|
||||||
| NC score | 0.015617 (rank : 71) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
GRB7_HUMAN
|
||||||
| NC score | 0.015278 (rank : 72) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14451, Q92568, Q96DF9 | Gene names | GRB7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth factor receptor-bound protein 7 (GRB7 adapter protein) (Epidermal growth factor receptor GRB-7) (B47). | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.011821 (rank : 73) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.010271 (rank : 74) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
ARBK1_HUMAN
|
||||||
| NC score | 0.009951 (rank : 75) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P25098, Q13837 | Gene names | ADRBK1, BARK, BARK1, GRK2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adrenergic receptor kinase 1 (EC 2.7.11.15) (Beta-ARK-1) (G- protein coupled receptor kinase 2). | |||||
|
MORN1_HUMAN
|
||||||
| NC score | 0.008677 (rank : 76) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
ARBK1_MOUSE
|
||||||
| NC score | 0.008004 (rank : 77) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99MK8, Q99LL8 | Gene names | Adrbk1, Grk2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adrenergic receptor kinase 1 (EC 2.7.11.15) (Beta-ARK-1) (G- protein-coupled receptor kinase 2). | |||||
|
ARAF_HUMAN
|
||||||
| NC score | 0.003650 (rank : 78) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||